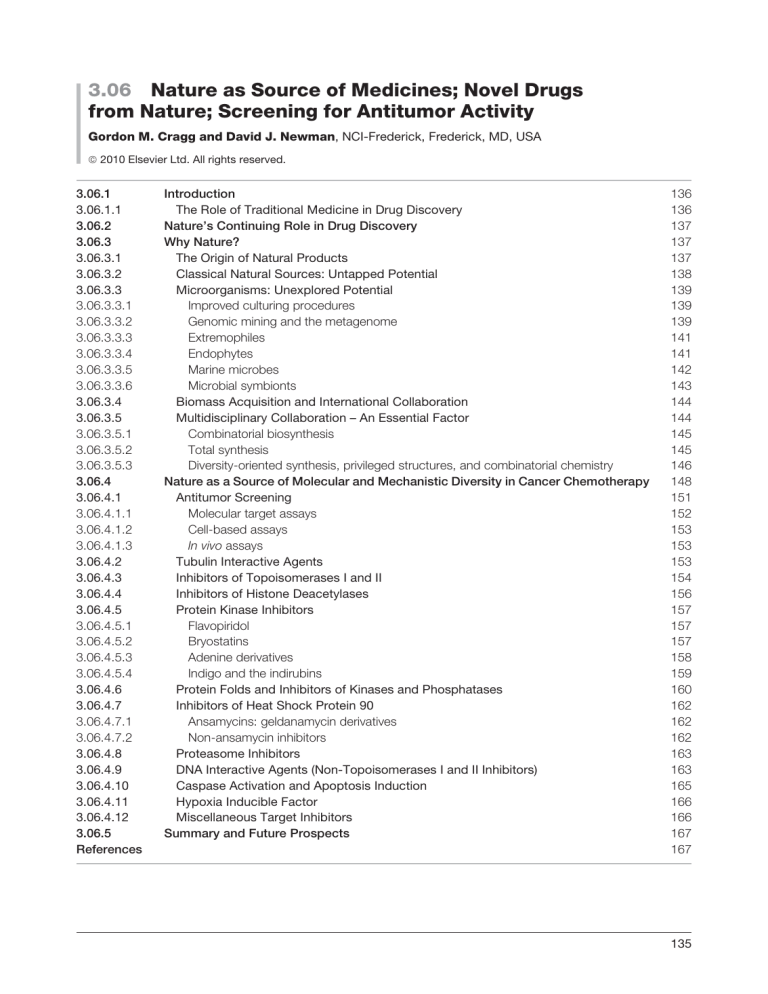
3.06 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Gordon M. Cragg and David J. Newman, NCI-Frederick, Frederick, MD, USA ª 2010 Elsevier Ltd. All rights reserved. 3.06.1 3.06.1.1 3.06.2 3.06.3 3.06.3.1 3.06.3.2 3.06.3.3 3.06.3.3.1 3.06.3.3.2 3.06.3.3.3 3.06.3.3.4 3.06.3.3.5 3.06.3.3.6 3.06.3.4 3.06.3.5 3.06.3.5.1 3.06.3.5.2 3.06.3.5.3 3.06.4 3.06.4.1 3.06.4.1.1 3.06.4.1.2 3.06.4.1.3 3.06.4.2 3.06.4.3 3.06.4.4 3.06.4.5 3.06.4.5.1 3.06.4.5.2 3.06.4.5.3 3.06.4.5.4 3.06.4.6 3.06.4.7 3.06.4.7.1 3.06.4.7.2 3.06.4.8 3.06.4.9 3.06.4.10 3.06.4.11 3.06.4.12 3.06.5 References Introduction The Role of Traditional Medicine in Drug Discovery Nature’s Continuing Role in Drug Discovery Why Nature? The Origin of Natural Products Classical Natural Sources: Untapped Potential Microorganisms: Unexplored Potential Improved culturing procedures Genomic mining and the metagenome Extremophiles Endophytes Marine microbes Microbial symbionts Biomass Acquisition and International Collaboration Multidisciplinary Collaboration – An Essential Factor Combinatorial biosynthesis Total synthesis Diversity-oriented synthesis, privileged structures, and combinatorial chemistry Nature as a Source of Molecular and Mechanistic Diversity in Cancer Chemotherapy Antitumor Screening Molecular target assays Cell-based assays In vivo assays Tubulin Interactive Agents Inhibitors of Topoisomerases I and II Inhibitors of Histone Deacetylases Protein Kinase Inhibitors Flavopiridol Bryostatins Adenine derivatives Indigo and the indirubins Protein Folds and Inhibitors of Kinases and Phosphatases Inhibitors of Heat Shock Protein 90 Ansamycins: geldanamycin derivatives Non-ansamycin inhibitors Proteasome Inhibitors DNA Interactive Agents (Non-Topoisomerases I and II Inhibitors) Caspase Activation and Apoptosis Induction Hypoxia Inducible Factor Miscellaneous Target Inhibitors Summary and Future Prospects 136 136 137 137 137 138 139 139 139 141 141 142 143 144 144 145 145 146 148 151 152 153 153 153 154 156 157 157 157 158 159 160 162 162 162 163 163 165 166 166 167 167 135 136 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 3.06.1 Introduction Throughout the ages humans have relied on nature to cater for their basic needs, not the least of which are medicines for the treatment of a wide spectrum of diseases. Plants, in particular, have formed the basis of sophisticated traditional medicine systems, with the earliest records, dating from around 2600 BCE, documenting the uses of approximately 1000 plant-derived substances in Mesopotamia, many of which are still used today for the treatment of ailments ranging from coughs and colds to parasitic infections and inflammation.1 Egyptian medicine dates from about 2900 BC, but the best-known record is the ‘Ebers Papyrus’ dating from 1500 BCE, documenting over 700 drugs, mostly of plant origin. The Chinese materia medica has been extensively documented over the centuries,2 with the first record dating from about 1100 BCE (Wu Shi Er Bing Fang, containing 52 prescriptions), followed by works such as the Shennong Herbal (100 BCE; 365 drugs) and the Tang Herbal (CE 659; 850 drugs). Likewise, documentation of the Indian ayurvedic system dates from before 1000 BCE (Charaka; Sushruta and Samhitas with 341 and 516 drugs, respectively).3,4 The Greeks and Romans contributed substantially to the rational development of the use of herbal drugs in the ancient Western world. Dioscorides, a Greek physician (CE 100), accurately recorded the collection, storage, and use of medicinal herbs during his travels with Roman armies throughout the then ‘known world’, while Galen (CE 130–200), a practitioner and teacher of pharmacy and medicine in Rome, was well known for his complex prescriptions and formulae used in compounding drugs. The Arabs, however, preserved much of the Greco-Roman expertise during the Dark and Middle Ages (fifth to twelfth centuries), and expanded it to include the use of their own resources, together with Chinese and Indian herbs unknown to the Greco-Roman world. A comprehensive review of the history of medicine may be found on the website of the National Library of Medicine (NLM), US National Institutes of Health (NIH), at http://www.nlm.nih.gov/hmd/index.html. 3.06.1.1 The Role of Traditional Medicine in Drug Discovery Plant-based systems continue to play an essential role in health care, and their use by different cultures has been extensively documented.5,6 The World Health Organization (WHO) has estimated that approximately 65% of the world’s population rely mainly on plant-derived traditional medicines for their primary health care, while plant products also play an important role in the health care systems of the remaining population, mainly residing in developed countries.7 A survey of plant-derived pure compounds used as drugs in countries hosting WHO-Traditional Medicine Centers indicated that, of 122 compounds identified, 80% were used for the same or related ethnomedical purposes and were derived from only 94 plant species.7,8 Probably the best example of ethnomedicine’s role in guiding drug discovery and development is that of the antimalarial drugs, particularly quinine and artemisinin. Malaria remains one of the greatest health challenges confronting humankind, and the search for better drugs, both in terms of efficacy and cost, is a global health imperative. The isolation of the antimalarial drug, quinine, from the bark of Cinchona species (e.g., Cinchona officinalis), was reported in 1820 by the French pharmacists, Caventou and Pelletier.9 The bark had long been used by indigenous groups in the Amazon region for the treatment of fevers and was first introduced into Europe in the early 1600s for the treatment of malaria. Quinine formed the basis for the synthesis of the commonly used antimalarial drugs, chloroquine and mefloquine, which largely replaced quinine in the mid-twentieth century, but with the emergence of resistance to both these drugs in many tropical regions, another plant long used in the treatment of fevers in traditional Chinese medicine (TCM), Artemisia annua (Quinhaosu), gained prominence.10 The discovery of artemisinin (1; Figure 1) by Chinese scientists in 1971 provided an exciting new natural product lead compound,11 and artemisinin analogues are now used for the treatment of malaria in many countries.12 Many analogues of artemisinin have been prepared in attempts to improve its activity and utility,12 and two of the most promising of these are the totally synthetic analogue OZ277 (2; Figure 1),13 and the dimeric analogue (3; Figure 1). Single doses of the latter compound were shown to cure malaria-infected mice, while corresponding treatments with artemisinin were much less effective.14 Although plants have a long history of use in the treatment of cancer,15 many of the claims for the efficacy of such treatment should be viewed with some skepticism because cancer, as a specific disease entity, is likely to be poorly defined in terms of folklore and traditional medicine.16 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 137 Figure 1 Natural antimalarial agents and analogues. 3.06.2 Nature’s Continuing Role in Drug Discovery The authors have reviewed the continuing valuable contributions of nature as a source of potential chemotherapeutic agents.17 In our paper, we analyzed the sources of new drugs over the period from January 1981 to June 2006, and classified these compounds as N (an unmodified natural product); ND (a modified natural product); S (a synthetic compound with no natural product conception); S ; S /NM (a synthetic compound with a natural product pharmacophore; S /NM indicating competitive inhibition); and S/NM (a synthetic compound showing competitive inhibition of the natural product substrate). This analysis indicated that while 66% of the 974 small molecules, new chemical entities (NCEs), are formally synthetic, 17% correspond to synthetic molecules containing pharmacophores derived directly from natural products classified as S and S/NM. Furthermore, 12% are actually modeled on a natural product inhibitor of the molecular target of interest, or mimic (i.e., competitively inhibit) the endogenous substrate of the active site, such as ATP (S/NM). Thus, only 37% of the 974 NCEs can be classified as truly synthetic (i.e., devoid of natural inspiration) in origin (S) (Figure 2). Considering disease categories, close to 70% of anti-infectives (antibacterial, antifungal, antiparasitic, and antiviral) are naturally derived or inspired (N; ND; S ; S /NM; S/NM), while in the cancer treatment area 77.8% are in this category, with the figure being 63% if the S/NM category is excluded. 3.06.3 Why Nature? 3.06.3.1 The Origin of Natural Products While the contributions of natural secondary metabolites to modern medicine are abundantly clear, the reasons for the production of these inherently biologically active compounds by organisms are still debated. Initially they were regarded as waste products, but it seems reasonable to assume that, in many instances, the production S* 5% N 6% S*/NM 12% ND 28% S/NM 12% S 37% Figure 2 Small molecule new chemical entities, January 1981 to June 2006, by source (N ¼ 974). 138 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 3 Secondary metabolites in chemical defense and quorum sensing. of these complex and often toxic chemicals has evolved over aeons as a means of chemical defense by essentially stationary organisms such as plants and many marine invertebrates, against predation and consumption (e.g., herbivory). For instance, pupae of the coccinellid beetle, Epilachna borealis, appear to exert a chemical defensive mechanism against predators through the secretion of droplets from their glandular hairs containing a library of hundreds of large-ring (up to 98 members) macrocyclic polyamines, with the simplest example having the generic formula shown (4; Figure 3).18 These libraries are built up from three simple 2-hydroxyethylamino-alkanoic acid precursors and are clear evidence that combinatorial chemistry has been pioneered and widely used in nature for the synthesis of biologically active compound libraries. Microorganisms are reported to produce and excrete antimicrobial toxins as a means of killing sensitive strains of the same or related species.19 This is similar to allelopathy in which plants release toxic compounds in order to suppress the growth of neighboring plants.20,21 Bacteria also control their density of population growth and the so-called biofilm formation through a cell-to-cell signaling mechanism known as quorum sensing involving the excretion of quorum-sensing compounds. The best studied of these are the acyl homoserine lactones (AHLs), with the compounds from Vibrio fischeri being examples; these include N-3-oxohexanoyll-homoserine lactone (5; Figure 3) and a previously unidentified furanone boronate diester that appears to be a universal signal (6; Figure 3), and they signal the activation of genes promoting virulence, spore formation, biofilm formation, and other phenomena.22,23 Natural products may be used for purposes of both predation and defense. Thus, species of the cone snail genus, Conus, stun their prey before capturing by the injection of venom composed of combinatorial libraries of several hundred peptides24 and the venom may also be used for defense against predators. One component of this mixture has been developed as Ziconotide, a nonnarcotic analgesic that is currently marketed as Prialt.25 3.06.3.2 Classical Natural Sources: Untapped Potential Despite the intensive investigation of terrestrial flora, it is estimated that only 6% of the approximately 300 000 species (some estimates are as high as 500 000 species) of higher plants have been systematically investigated, pharmacologically, and only some 15% phytochemically.8,26,27 The potential of the marine environment as a source of novel drugs remains virtually unexplored,28,29 and until recently, the investigation of the marine environment had largely been restricted to tropical and subtropical regions; however, the exploration is being expanded to colder regions. The isolation of novel pyrido-pyrrolo-pyrimidine derivatives, the variolins (e.g., variolin B: 7; Figure 4), from the Antarctic sponge, Kirkpatrickia variolosa, was reported in 1994,30,31 followed by their total synthesis in 2003,32 while the isolation of a cytotoxic macrolide palmerolide A (8; Figure 4) from an Antarctic tunicate has recently been reported,33 with total synthesis leading to a revision of the original structure.34 The selective and reproducible production of bioactive compounds has been induced through exposure of the roots of hydroponically grown plants to chemical elicitors,35 while feeding of seedlings with derivatives of selected biosynthetic precursors can lead to the production of nonnatural analogues of the natural metabolites. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 139 Figure 4 Natural products from novel sources. Thus, the production of nonnatural terpene indole alkaloids related to the vinca alkaloids has been reported through the feeding of seedlings of Catharanthus roseus with various tryptamine analogues.36 3.06.3.3 Microorganisms: Unexplored Potential Until recently, the inability to cultivate most naturally occurring microorganisms has severely limited the study of natural microbial ecosystems, and it has been estimated that much less than 1% of microorganisms seen microscopically have been cultivated. Yet, despite this limitation, the number of highly effective microbially derived chemotherapeutic agents discovered and developed thus far has been highly impressive. Given the observation that ‘‘a handful of soil contains billions of microbial organisms,’’37 and the assertion that ‘‘the workings of the biosphere depend absolutely on the activities of the microbial world,’’38 the microbial universe clearly presents a vast untapped resource for drug discovery. In addition, substantial advances in the understanding of the gene clusters encoding multimodular enzymes involved in the biosynthesis of a multitude of microbial secondary metabolites, such as polyketide synthases (PKSs) and/or nonribosomal peptide synthetases (NRPSs), has enabled the sequencing and detailed analysis of the genomes of long-studied microbes such as Streptomyces avermitilis. These studies have revealed the presence of additional PKS and NRPS clusters resulting in the discovery of novel secondary metabolites not detected in standard fermentation and isolation processes.39 Such genome mining has been used in the discovery of a novel peptide, coelichelin, from the soil bacterium, Streptomyces coelicolor,40 and this concept is further expanded in the discussion in Section 3.06.3.3.2. 3.06.3.3.1 Improved culturing procedures Recent developments of procedures for cultivating and identifying microorganisms are aiding microbiologists in their assessment of the earth’s full range of microbial diversity. For example, the use of ‘nutrient-sparse’ media simulating the original natural environment enables the massive parallel cultivation of gel-encapsulated single cells (gel microdroplets (GMDs)) derived from microbes separated from environmental samples (seawater and soil).41 This has permitted ‘‘the simultaneous and relatively noncompetitive growth of both slow- and fast-growing microorganisms,’’ thereby preventing the overgrowth by fast-growing ‘microbial weeds’, and leading to the identification of previously undetected species (using 16S rRNA gene sequencing), as well as the culturing and scale-up cultivation of previously uncultivated microbes. Coupled with the recent report of the sequencing of the marine actinomycete, Salinospora tropica, where it was found that approximately 10% of the genome coded for potential secondary metabolites,42 and the recent paper on cultivation of Gram-positive marine microbes,43 the potential for discovery of novel agents is immense. 3.06.3.3.2 Genomic mining and the metagenome Despite improvement in culturing techniques, greater than 99% of microscopically observed microbes still defy culture. Extraction of nucleic acids (the metagenome) from environmental samples, however, permits the identification of uncultured microorganisms through the isolation and sequencing of ribosomal RNA or rDNA (genes encoding for rRNA). Samples from soils and seawater are currently being investigated,44,45 and whole-genome shotgun sequencing of environmental-pooled DNA obtained from water samples collected in the Sargasso Sea off 140 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity the coast of Bermuda by the Venter group, indicated the presence of at least 1800 genomic species, which included 148 previously unknown bacterial phylotypes.45 Venter and his coworkers46 are also examining microbial communities in water samples collected by the Sorcerer II Global Ocean Sampling (GOS) expedition, and their data predict more than six million proteins, nearly twice the number of proteins present in current databases, with some of the predicted proteins bearing no similarity to any currently known proteins, and therefore representing new families. These methods may be applied to other habitats, such as the microflora of insects47 and marine animals,48 and there is a recent report of an ‘Air Genome Project’ being launched in Manhattan where samples of air are being analyzed for the content of DNA from bacteria, fungi, and other microbes.49 The cloning and understanding of the novel genes discovered through these processes, and the heterologous expression of gene clusters encoding the enzymes involved in biosynthetic pathways in viable host organisms, such as Escherichia coli, should permit the production of novel metabolites produced from as yet uncultured microbes. The enormous unexplored potential of microbial diversity and the strategy of genome mining were briefly mentioned in the introduction of Section 3.06.3.3. As a result of the rapid evolution of genomic sequencing and the ever-dropping costs of performing such studies, the amount of genomic information is ever increasing, resulting in the potential for the expression of previously unrecognized metabolites. A recent review discusses the general aspects of genomics in natural product research.50 It has now become evident, initially through the pioneering work of Hopwood, that the genome of the Streptomycetes and by extension, Actinomycetes in general, contain large numbers of previously unrecognized secondary metabolite clusters. An excellent example is the investigation of the genome of the well-known vancomycin producer, Amycolatopsis orientalis (ATCC 43491), which resulted in the isolation of the novel antibiotic ECO-0501 (9; Figure 5), which was only found by using the genomic sequence to predict the molecular weight, and then looking for the molecule directly by high performance liquid chromatography–mass spectrometry (HPLC–MS). The compound had a very similar biological profile to vancomycin but was masked by this compound.51 Many more examples of the value of this type of investigation have been provided in two recent reviews,52,53 which give up-to-date information on the manifold structures that can be found by expression of environmental DNA. Figure 5 New compounds from genome mining. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 141 The presence of potential gene products controlling metabolite production has been predicted in a recently reported genomic analysis of the fungus Aspergillus nidulans, which not only suggested the presence of clustered secondary metabolite genes having the potential to generate up to 27 polyketides, 14 nonribosomal peptides (NRPs), 1 terpene, and 2 indole alkaloids, but also identified the potential controller of expression of these clusters.54 This was demonstrated by expressing terrequinone A (10; Figure 5), a compound not previously reported from this species.54 Similar predictions can be made from Aspergillus fumigatus and Aspergillus oryzae as a result of the analysis of the potential number of secondary metabolite clusters in these fungi.54 A recent review expands the discussion on control of secondary metabolites in fungi.55 Even the myxobacteria have now yielded to genomic analyses, and the identification and utilization of ChiR, the gene-controlling production of chivosazol (11; Figure 5), an extremely potent eukaryotic antibiotic, has been reported.56 This paper also deals with the major problem in secondary metabolite expression, whether in homologous or heterologous hosts, which is the identification and application of the transcriptional control mechanisms involved. 3.06.3.3.3 Extremophiles Extremophilic microbes (extremophiles) abound in extreme habitats. These include acidophiles (acidic sulfurous hot springs), alkalophiles (alkaline lakes), halophiles (salt lakes), piezo (baro-) and (hyper)thermophiles (deep-sea vents),57–61 and psychrophiles (Arctic and Antarctic waters, alpine lakes).62 Thus far, investigations have centered on the isolation of thermophilic and hyperthermophilic enzymes (extremozymes),63–67 but there is little doubt that these extreme environments will also yield novel bioactive chemotypes. Abandoned mine-waste disposal sites have yielded unusual acidophiles, which thrive in the acidic, metal-rich waters, polluted environments that are generally toxic to most prokaryotic and eukaryotic organisms.68 The novel sesquiterpenoid and polyketide-terpenoid metabolites, berkeleydione (12; Figure 6) and berkeleytrione (13; Figure 6) showing activity against metalloproteinase-3 and caspase-1, activities relevant to cancer, Huntington’s disease, and other diseases, have been isolated from Penicillium species found in the surface waters of Berkeley Pit Lake in Montana.68–70 3.06.3.3.4 Endophytes As indicated in Sections 3.06.1 and 3.06.3.2, plants have been relatively extensively studied as sources of bioactive metabolites, but the endophytic microbes that reside in the tissues between living plant cells have received little attention. Relationships between endophytes and their host plants may vary from symbiotic to pathogenic, and studies have revealed an interesting realm of novel chemistry.71–73 A wide range of new bioactive molecules have been discovered, including peptide antibiotics, the coronamycins, isolated from a Streptomyces species associated with an epiphytic vine (Monastera species) found in the Peruvian Amazon,74 and the cytotoxic aspochalasins I, J, and K (14–16, respectively; Figure 7), isolated from endophytes of plants from the southwestern desert regions of the United States.75 Of particular significance has been the production of various important anticancer agents in small quantities from endophytic fungi isolated from plants. Examples (Figure 7) are Taxol (17) from Taxomyces76 and many Pestalotiopsis species,77 as well as camptothecin (18),78,79 podophyllotoxin, an epimer of the precursor to the anticancer drug, etoposide (19),80,81 vinblastine (20),82 and vincristine (21)83,84 from endophytic fungi isolated Figure 6 New compounds from extreme environments. 142 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 7 Natural products from endophytes. from the original source plants. It has been demonstrated that these compounds are not artifacts, and so the identification of the gene/gene product controlling metabolite production by these microbes could provide an entry into greatly increased production of key bioactive natural products. 3.06.3.3.5 Marine microbes Deep-ocean sediments are proving to be a valuable source of new actinomycete bacteria that are unique to the marine environment.42 Use of a combination of culture and phylogenetic approaches has led to the description of the first truly marine actinomycete genus named Salinospora,43,85 and its members are proving to be ubiquitous, being found in concentrations of up to 104 per milliliter in sediments on tropical ocean bottoms and in more shallow waters, as well as appearing on the surfaces of numerous marine plants and animals. On culturing using the appropriate selective isolation techniques, significant antibiotic and cytotoxic activity has been observed, and has resulted in the isolation of a potent cytotoxin, salinosporamide A (22; Figure 8), a very potent proteasome inhibitor (IC50 ¼ 1.3 nmol l1),86 currently in Phase I clinical trials. More recently, the isolation and cultivation of another new actinomycete genus, named Marinispora, has been reported, and novel macrolides called marinomycins have been isolated. Marinomycins A (23; Figure 8) to D show potent activity against drug-resistant bacterial pathogens and some melanomas.87 Recent publications by the Fenical group on the novel and diverse chemistry of these new microbial genera include the isolation of potential chemopreventive agents, saliniketals A and B from Salinispora arenicola,88 while two new cyclic peptides, thalassospiramides A and B, possessing immunosuppressive activity have been isolated from a new member of the marine -proteobacterium Thalassospira.89 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 143 Figure 8 Examples of novel microbial natural products. 3.06.3.3.6 Microbial symbionts Evidence is mounting indicating that many bioactive compounds isolated from various macroorganisms are actually metabolites synthesized by symbiotic bacteria.90 These include the anticancer compounds, the maytansanoids (24; Figure 8), originally isolated from several plant genera of the Celastraceae family,91 and the pederins (25; Figure 8), isolated from beetles of genera Paederus and Paederidus as well as from several marine sponges.92–94 In addition, a range of antitumor agents isolated from marine organisms closely resemble bacterial metabolites.28 An interesting example of a complex symbiotic–pathogenic relationship involving a bacterium–fungus– plant interaction has been discovered in the case of rice seedling blight. The toxic metabolite, rhizoxin (26; Figure 8), originally isolated from the contaminating Rhizopus fungus, has actually been found to be produced by an endosymbiotic Burkholderia bacterial species.95 Rhizoxin exhibits potent antitumor activity, but its further development as an anticancer drug has been precluded by toxicity problems. Thus, in addition to 144 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity offering potentially new avenues for pest control, this unexpected finding has enabled the isolation of rhizoxin as well as rhizoxin analogues through the cultivation of the bacterium independently of the fungal host. This may have significant implications in the development of agents with improved pharmacological properties. 3.06.3.4 Biomass Acquisition and International Collaboration The acquisition of biomass has changed significantly from the days when drug companies and others routinely collected organisms with little thought of ownership by, or benefit sharing with, the source country. Today, thanks to the Convention on Biodiversity (CBD) and similar documents and agreements such as the US National Cancer Institute’s (NCI) Letter of Collection (LOC: http://ttc.nci.nih.gov/forms/), all ethical biomass acquisitions now include provisions for the source country to be compensated in some way for the use of its biomass. It should be noted that the LOC predated the CBD by 3 years; its terms, as a minimum, must be adhered to by any investigator who has his or her collections funded by the NCI/NIH. Such recompense to the source country is best provided through formal agreements with government organizations and collectors in the source country, with such agreements providing not only for reimbursement of collecting expenses but also for further benefits (often in the form of milestone and/or royalty payments) in the event that a drug is developed from a collected sample. Agreements often include terms related to the training of source country scientists and transfer of technologies involved in the early drug discovery process. Recognition of the role played by indigenous peoples through the stewardship of resources in their region and/or the sharing of their ethnopharmacological information in guiding the selection of materials for collection is important in determining the distribution of such benefits. There have been sample legal agreements96 and discussions as to methods used by various groups published in the last few years.97–99 All samples collected, irrespective of type of source, must if at all possible be fully identified to genus and species. Such identification is usually possible for all plant species, but it is not always possible for microbes and marine organisms. Voucher specimens should be provided to an appropriate depository in the source country as well as to a similar operation in the home country of the collector. The use of traditional knowledge in the drug discovery process has been briefly discussed in Section 3.06.1.1. The selection of plant samples using such knowledge, the ethnobotanical/ethnopharmacological approach, usually involves the selection of plants that have a documented (written or oral) use by traditional healers and has the advantage of tapping into the empirical knowledge developed over centuries of use by large numbers of people. In addition, the bioactive constituents may be considered as having had a form of continuing clinical trial in man. The benefits of this approach have been extolled in several relatively recent articles,100–102 but a weakness of the ethnobotanical approach has always been that it is slow, requiring careful interviewing of traditional healers by skilled scientists, including ethnobotanists, anthropologists, trained physicians, and pharmacologists. In addition, the quoted medicinal activity in the collected plant(s) may not be detectable, given the particular assays used by the screening laboratory. The highest possibility of success of the ethnobotanical approaches lies in studies related to overt diseases/conditions such as parasitic infections, fungal sores, and contraception and conception to name a few. In such cases, there are adequate controls, even on the same patient. Where there does not yet appear to be any successful relationship is in diseases such as cancer and AIDS-related conditions, where extensive testing of the patient is required for an accurate diagnosis.16 3.06.3.5 Multidisciplinary Collaboration – An Essential Factor The probability that a directly isolated natural product (e.g., adriamycin or taxanes in the antitumor area) will be the drug used for the treatment of a given disease in the future is relatively low. In many instances, however, these natural molecules can serve as lead compounds that can be optimized through the application of methodologies such as combinatorial biosynthesis and/or combinatorial chemistry to give products suitable for drug development. In addition, novel methods of total chemical syntheses of the natural molecules can yield intermediates possessing equal or superior preclinical activity to that observed for the natural product, and that can be optimized for drug development using medicinal or combinatorial chemistry approaches. Of course, all these approaches require suitable biological assays for evaluation of the optimization products, and thus a truly Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 145 multidisciplinary, collaborative approach is required for effective drug development. That these ideas are not just pipe dreams can be seen in the following examples. 3.06.3.5.1 Combinatorial biosynthesis The substantial advances made in the understanding of the role of multifunctional PKS enzymes in bacterial aromatic polyketide biosynthesis have led to the identification of many such enzymes, together with their encoding genes.103–106 The same applies to NRPSs responsible for the biosynthesis of NRPs.105 The rapid developments in the analysis of microbial genomes have enabled the identification of a multitude of gene clusters encoding for polyketides, NRPs, and hybrid polyketide–NRP metabolites, and have provided the tools for engineering the biosynthesis of novel ‘nonnatural’ natural products through gene shuffling, domain deletions, and mutations.105,107 Results of the application of these combinatorial biosynthetic techniques to the production of novel analogues of anticancer agents, such as the anthracyclines, ansamitocins, epothilones, enediynes, and aminocoumarins, have recently been reviewed by Shen et al.108 The efficient scale-up production of epothilone D exemplifies the power of this technique. Epothilone D, the des-epoxy precursor of epothilone B, was the most active of the epothilone series isolated from the myxobacterium, Sorangium cellulosum, and entered clinical trials as a potential anticancer agent but has now been discontinued in favor of a congener, 9,10-didehydroepothilone D. The polyketide gene cluster producing epothilone B has been isolated and sequenced from two S. cellulosum strains,109,110 and the epoxidation of epothilone D to epothilone B has been shown to be due to the last gene in the cluster, epoK, encoding a cytochrome P-450. Heterologous expression of the gene cluster minus the epoK in Myxococcus xanthus resulted in large-scale production of crystalline epothilone D.111 3.06.3.5.2 Total synthesis The total synthesis of complex natural products has long posed challenges to the top synthetic chemistry groups worldwide and has led to dramatic advances in the field of organic chemistry.112 As eloquently stated by Nicolaou and his coauthors, ‘‘Today, natural product total synthesis is associated with prudent and tasteful selection of challenging and preferably biologically important target molecules; the discovery and invention of new synthetic strategies and technologies; and explorations in chemical biology through molecular design and mechanistic studies. Future strides in the field are likely to be aided by advances in the isolation and characterization of novel molecular targets from nature, the availability of new reagents and synthetic methods, and information and automation technologies.’’112 In some instances, as noted in Section 3.06.3.2 regarding the cytotoxic macrolide palmerolide A (8; Figure 4), total synthesis has led to a revision of the original published structure;34 another notable example is that of the marine-derived antitumor compound, diazonamide A (27; Figure 9).113 Significant strides have been made in the synthesis and structural modification of drugs that are difficult to isolate in sufficient quantities for development. Adequate supply can be a serious limiting factor in the preclinical and clinical development of some naturally derived drugs, and the focus of many top synthetic groups on devising economically feasible synthetic strategies is a very welcome development for both clinicians conducting clinical trials and patient populations. An excellent example is the marine-derived anticancer agent discodermolide (28; Figure 9), where total synthesis provided sufficient quantities for thorough clinical trials, but unfortunately, these have now been terminated due to the lack of objective responses and toxicity.114,115 The process of total synthesis can often lead to the identification of a substructural portion of the molecule bearing the essential features necessary for activity (the pharmacophore), and, in some instances, this has resulted in the synthesis of simpler analogues having similar or better activity than the natural product itself. One of the most notable examples is that of the marine-derived antitumor agent, halichondrin B (29; Figure 9), where total synthetic studies revealed that the right-hand half of the molecule retained all or most of the potency of the parent compound, and the analogue, E7389 (Eribulin) (30; Figure 9), is currently in Phase III clinical trials.116 In some instances, clinical trials of the original natural product may fail, but totally synthetic analogues continue to be developed. Thus, while clinical trials of the marine-derived anticancer agents, dolastatin 10 and dolastatin 15, have been terminated, the synthetic analogues of dolastatin 10 (31; Figure 9), TZT-1027 (auristatin PE or soblidotin) and ILX651 (synthadotin or tasidotin) based on dolastatin 15, are in Phase II clinical trials.117 146 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 9 Products of total synthesis. 3.06.3.5.3 Diversity-oriented synthesis, privileged structures, and combinatorial chemistry While there are claims that combinatorial chemistry is generating new leads,118 the declining numbers of new NCEs119 indicate that the use of de novo combinatorial chemistry approaches to drug discovery over the past decade have been disappointing, with some of the earlier libraries being described as ‘‘poorly designed, impractically large, and structurally simplistic.’’118 As stated in this article, ‘‘an initial emphasis on creating mixtures of very large numbers of compounds has largely given way in industry to a more measured approach based on arrays of fewer, well-characterized compounds’’ with ‘‘a particularly strong move toward the synthesis of complex natural product-like compounds – molecules that bear a close structural resemblance to approved natural product-based drugs.’’ The importance of the use of natural product-like scaffolds for generating meaningful combinatorial libraries has been further emphasized in a recent article entitled ‘‘Rescuing Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 147 Combichem diversity-oriented synthesis (DOS) aims to pick up where traditional combinatorial chemistry left off.’’120 In this article it is stated that ‘‘the natural product-like compounds produced in DOS have a much better shot at interacting with the desired molecular targets and exhibiting interesting biological activity.’’ The synthesis of natural product-like libraries is exemplified by the work of the Schreiber group who have combined the simultaneous reaction of maximal combinations of sets of natural product-like core structures (latent intermediates) with peripheral groups (skeletal information elements) in the synthesis of libraries of over 1000 compounds bearing significant structural and chiral diversity.121,122 Through detailed analyses of active natural product skeletons, relatively simple key precursor molecules may be identified, which form the building blocks for use in combinatorial synthetic schemes, thereby enabling structure–activity relationships (SARs) to be probed. The generation of small libraries, built through the solid-phase synthesis of molecules such as epothilone A (32; Figure 10), dysidiolide (33; Figure 10), galanthamine (34; Figure 10), and psammaplin (35; Figure 10), has been reviewed.123–129 Use of an active natural product as the central scaffold can also be applied to the generation of large numbers of analogues for structure–activity studies, the so-called parallel synthetic approach, and is exemplified by the syntheses around the sarcodictyin (36; Figure 10) scaffold.130 The importance of natural products as leads for combinatorial synthesis is further illustrated by the concept of ‘‘privileged structures,’’131 and this approach has been successfully developed by several groups.129,132–134 In one such case, a search of the literature yielded nearly 4000 2,2-dimethyl-2H-benzopyran moieties (37; Figure 10), with another 8000 structures identified Figure 10 Diversity-oriented and parallel synthesis and privileged structures. 148 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity through the inclusion of a slight modification of the search. Application of solid-phase synthetic methods led to the identification and subsequent optimization of benzopyrans with a cyanostilbene substitution (38; Figure 10) that are effective against vancomycin-resistance bacteria.132–134 3.06.4 Nature as a Source of Molecular and Mechanistic Diversity in Cancer Chemotherapy A list of all anticancer drugs currently in clinical use and classified according to their source is given in Table 1. Readers are referred to the authors’ 2007 review17 for detailed references. In the sections below, after briefly reviewing the various methodologies used in antitumor screening, we have provided an overview of the chemotherapeutic agents currently in clinical use or development for the treatment of cancer. Our discussion of these agents is divided into subsections based on their mechanisms of action. The discussions are brief since other chapters in this series will be dealing in more detail with many of the agents mentioned. Information on ongoing clinical trials may be found at http://www.clinicaltrials.gov/, and readers are referred to this site for details. Table 1 All anticancer drugs (1940s to December 2007) (organized alphabetically by generic name within source) Generic name Year introduced Referencea 131I-chTNT H-101 Aldesleukin Alemtuzumab Bevacizumab Celmoleukin Cetuximab Denileukin diftitox Interferon alfa2a Interferon alfa2b Interferon, gamma-1a Interleukin-2 Mobenakin Nimotuzumab Panitumumab Pegaspargase Rexin-gb Rituximab Tasonermin Teceleukin Tositumomab Trastuzumab Aclarubicin Actinomycin D Angiotensin II Arglabin Asparaginase Bleomycin Carzinophilin Chromomycin A3 Daunomycin Doxorubicin Leucovorin Masoprocol Mithramycin 2007 2005 1992 2001 2004 1992 2003 1999 1986 1986 1992 1989 1999 2006 2006 1994 2007 1997 1999 1992 2003 1998 1981 1964 1994 1999 1969 1966 1954 1961 1967 1966 1950 1992 1961 I 393351 DNP 19 ARMC 25 DNP 15 ARMC 40 DNP 06 ARMC 39 ARMC 35 I 204503 I 165805 ARMC 28 ARMC 25 ARMC 35 DNP 20 DNP 20 ARMC 30 I 34631 DNP 11 ARMC 35 DNP 06 ARMC 39 DNP 12 I 090013 FDA ARMC 30 ARMC 35 FDA FDA Japan Antibiotics Japan Antibiotics FDA FDA FDA ARMC 28 FDA Page 46 314 38 450 102 346 338 332 314 345 29 28 306 25 349 102 364 35 296 335 333 Source B B B B B B B B B B B B B B B B B B B B B B N N N N N N N N N N N N N (Continued ) Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Table 1 (Continued) Generic name Year introduced Referencea Mitomycin C Neocarzinostatin Paclitaxel Palictaxel nanoparticlesc Paclitaxel nanoparticlesd Pentostatin Peplomycin Sarkomycin Solamargine (aka BEC) Trabectedin Streptozocin Testosterone Vinblastine Vincristine Kunecatechins Sinecatechins Alitretinoin Amrubicin hcl Belotecan hydrocholoride Calusterone Cladribine Cytarabine ocfosfate Dexamethasone Docetaxel Dromostanolone Elliptinium acetate Epirubicin HCI Estramustine Ethinyl estradiol Etoposide Exemestane Fluoxymesterone Formestane Fosfestrol Fulvestrant Gemtuzumab ozogamicin Goserelin acetate Hexyl aminolevulinate Histrelin Hydroxyprogesterone Idarubicin hydrochloride Irinotecan hydrochloride Ixabepilone Leuprolide Medroxyprogesterone acetate Megesterol acetate Methylprednisolone Methyltestosterone Miltefosine Mitobronitol Nadrolone phenylpropionate Norethindrone acetate Pirarubicin Prednisolone Prednisone Temsirolimus Teniposide Testolactone 1956 1976 1993 2005 2007 1992 1981 1954 1987 2007 Pre-1977 Pre-1970 1965 1963 2006 2007 1999 2002 2004 1973 1993 1993 1958 1995 1961 1983 1984 1980 Pre-1970 1980 1999 Pre-1970 1993 Pre-1977 2002 2000 1987 2004 2004 Pre-1970 1990 1994 2007 1984 1958 1971 1955 1974 1993 1979 1959 Pre-1977 1988 Pre-1977 Pre-1970 2007 1967 1969 FDA Japan Antibiotics ARMC 29 DNP 19 I 422122 ARMC 28 I 090889 FDA DNP 03 I 139221 FDA FDA DNP 20 I 283701 ARMC 35 ARMC 38 ARMC 40 FDA ARMC 29 ARMC 29 FDA ARMC 31 FDA I 091123 ARMC 20 FDA Page 342 45 334 25 24 333 349 449 335 335 341 318 FDA DNP 13 46 ARMC 29 337 ARMC 38 DNP 14 ARMC 23 I 300211 I 109865 357 23 336 ARMC 26 ARMC 30 I 293356 ARMC 20 FDA FDA FDA FDA ARMC 29 FDA FDA 303 301 ARMC 24 309 218793 FDA FDA 319 340 Source N N N N N N N N N N N N N N NBe NBe ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND (Continued ) 149 150 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Table 1 (Continued) Generic name Year introduced Referencea Page Source Topotecan hcl Triamcinolone Triptorelin Valrubicin Vapreotide acetate Vindesine Vinorelbine Zinostatin stimalamer Amsacrine Arsenic trioxide Bisantrene hydrochloride Busulfan Carboplatin Carmustine (BCNU) Chlorambucil Chlortrianisene Cis-diamminedichloroplatinum Cyclophosphamide Dacarbazine Diethylstilbestrol Flutamide Fotemustine Heptaplatin /SK-2053R Hexamethylmelamine Hydroxyurea Ifosfamide Lenalidomide Levamisole Lobaplatin Lomustine (CCNU) Lonidamine Mechlorethanamine Melphalan Mitotane Nedaplatin Nilutamide Nimustine hydrochloride Oxaliplatin Pamidronate Pipobroman Porfimer sodium Procarbazine Ranimustine Razoxane Semustine (MCCNU) Sobuzoxane Sorafenib mesylate Thiotepa Triethylenemelamine Zoledronic acid Anastrozole Bicalutamide Bortezomib Camostat mesylate Dasatinib Erlotinib hydrochloride Fadrozole hcl Gefitinib 1996 1958 1986 1999 2003 1979 1989 1994 1987 2000 1990 1954 1986 1977 1956 Pre-1981 1979 1957 1975 Pre-1970 1983 1989 1999 1979 1968 1976 2005 Pre-1981 1998 1976 1987 1958 1961 1970 1995 1987 Pre-1981 1996 1987 1966 1993 1969 1987 Pre-1977 Pre-1977 1994 2005 1959 Pre-1981 2000 1995 1995 2003 1985 2006 2004 1995 2002 ARMC 32 FDA I 090485 ARMC 35 I 135014 FDA ARMC 25 ARMC 30 ARMC 23 DNP 14 ARMC 26 FDA ARMC 22 FDA FDA BOYD FDA FDA FDA 320 ND ND ND ND ND ND ND ND S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S/NM S/NM S/NM S/NM S/NM S/NM S/NM S/NM 350 320 313 327 23 300 318 ARMC 19 ARMC 25 ARMC 35 FDA FDA FDA DNP 19 Boyd DNP 12 FDA ARMC 23 FDA FDA FDA ARMC 31 ARMC 23 Boyd ARMC 32 ARMC 23 FDA ARMC 29 FDA ARMC 23 318 313 348 ARMC 30 DNP 19 FDA Boyd DNP 14 ARMC 31 ARMC 31 ARMC 39 ARMC 21 DNP 20 ARMC 40 ARMC 31 ARMC 38 310 45 45 35 337 347 338 313 326 343 341 24 338 338 345 325 27 454 342 358 (Continued ) Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Table 1 151 (Continued) Generic name Year introduced Referencea Page Source Imatinib mesilate Lapatinib ditosylate Letrozole Nafoxidine Nilotinib hydrochloride Sunitinib maleate Tamoxifen Toremifene Aminoglutethimide Azacytidine Capecitabine Carmofur Clofarabine Cytosine arabinoside Decitabine Doxifluridine Enocitabine Floxuridine Fludarabine phosphate Fluorouracil Ftorafur Gemcitabine hcl Mercaptopurine Methotrexate Mitoxantrone HCI Nelarabine Thioguanine Uracil mustard Abarelix Bexarotene Pemetrexed Raltitrexed Tamibarotene Temozolomide Vorinostat Bcg live Hpv vaccine (Merck) Hpv vaccine (GSK) Melanoma theraccine 2001 2007 1996 Pre-1977 DNP 15 I 301036 ARMC 32 38 S/NM S/NM S/NM S/NM S/NM S/NM S/NM S/NM S S S S S S S S S S S S S S S S S S S S S /NM S /NM S /NM S /NM S /NM S /NM S /NM V V V V 2006 1973 1989 1981 Pre-1977 1998 1981 2005 1969 2006 1987 1983 1971 1991 1962 1972 1995 1953 1954 1984 2005 1966 1966 2004 2000 2004 1996 2005 1999 2006 1990 2006 2007 2001 I 386178 DNP 20 FDA ARMC 25 319 FDA ARMC 34 FDA DNP 19 FDA DNP 20 ARMC 23 ARMC 19 FDA ARMC 27 FDA FDA ARMC 31 FDA FDA ARMC 20 DNP 19 FDA FDA ARMC 40 DNP 14 ARMC 40 ARMC 32 DNP 19 ARMC 35 DNP 20 DNP 04 DNP 20 I 309201 DNP 15 311 27 319 44 27 332 318 327 344 321 45 446 23 463 315 45 350 27 104 26 38 a Refer to Newman and Cragg17 for decoding the reference citations in this column. No generic name; this is the trade name. c Abraxane (entirely different from below in particle source and approved in USA). d Nanoxel (entirely different from above in particle source and approved in India). e NB is a new classification (natural product/botanical); these agents are for genital warts but approved for sale with a disease indication. b 3.06.4.1 Antitumor Screening As mentioned in Section 3.06.3.5, the successful development of effective new drugs requires suitable assays to guide, not only the discovery of a bioactive lead but also the evaluation of products developed through optimization of the lead. Thus, a given organism provides the investigator with a complex library of unique bioactive constituents, analogous to the library of crude synthetic products initially produced by combinatorial chemistry techniques; the two approaches can be seen as complementary to each other, with each providing access to (initially) different lead structures. The task of the natural products researcher is to select those compounds of pharmacological interest through bioassay-guided fractionation of the ‘natural combinatorial 152 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity libraries’ produced by the extraction of organisms, and then to collaborate in the optimization and development of the lead natural product structure as discussed in Section 3.06.3.5 above. Fortunately, the means to do this efficiently are now at hand. 3.06.4.1.1 Molecular target assays Structural diversity is not the only reason why natural products are of interest to drug development. Natural products frequently possess highly selective and specific biological activities based on mechanisms of action. A striking illustration is the influence of natural products on many of the molecular processes operative in cell cycle progression, and details may be found at the website of the Roscoff Biological Station (http:// www.sb-roscoff.fr), which covers diagrams originally published by Meijer135 on natural products and the cell cycle, with a modified version shown in Figure 11. In the early days of natural products research, new compounds were simply isolated at random, or at best by the use of simple broad-based bioactivity screens based on antimicrobial or cytotoxic activities. Although these screens did result in the isolation of many bioactive compounds,136 they were considered to be too nonspecific for the next generation of drugs. Fortunately, a large number of robust and specific biochemical- and genetics-based screens using transformed cells, a key regulatory intermediate in a biochemical or genetic pathway, or a receptor–ligand interaction (often derived from the explosion in genomic information since the middle 1990s), are now in routine use. These screens will permit the more precise detection of bioactive compounds in the complex matrices that are natural product extracts. One interesting feature of such screens has increased the attractiveness of natural products to the pharmaceutical industry. The screens themselves are all highly automated and high-throughput (upward of 50 000 Trabectedin Nitrogen mustards Nitrosoureas Mitomycin C Hydroxyurea Cytarabine Antifolates 5-Fluorouracil 6-Mercaptopurine Wortmannin Caffeine Fumagillin,TNP-470 PRIMA-1, pifithrin α UCN-01, SB-218078 Debromohymenialdisine Isogranulatimide Menadione (K3) (R)-roscovitine (CYC202) Paullones, indirubins p53/MDM2 ATM/ATR Chk1 Nucleotide excision Chk2 Repair Vinca alkaloids PD0166285 Taxol/taxotere Halichondrin CDC25 HMGA Spongistatin FK317 S CDK1 Wee1 Rhizoxin Camptothecin Topoisomerase i Aurora Cryptophycin Pin1 Tubulin Podophyllotoxin,doxorubicin Topoisomerase ii Sarcodictyin M Polymerization/ Etoposide, mitoxantrone CDK2 Eleutherobin depolymerization Epothilones Cdc7 (R)-Roscovitine (CYC 202) Discodermolide CDK4 Kinesin Eg5 Paullones, indirubins Indibulin ODC/SAMDC Actin G1 Dolastatin GSK-3 Flavopiridol Pin1 Combretastatin AhR Polyamine analogues Monastrol Eribulin MEK1/Erk - 1/2 G0 Cytochalasins Raf Paullones, indirubins ROCK Latrunculin A Farnesyl transferase DF203 Scytophycins Tyrosine kinases PD98059, U0126 Dolastatin 11 Proteasome PS-341 Jaspamide Sorafenib* Choline kinase CT-2584 Y-27632 mTOR/FRAP Rapamycin Tipifarnib Gleevec Bryostatin, PKC412 PKC Lonafarnib iressa HSP90 Geldanamycin, 17-AAG erlotinib Cytosolic phospholipase A2 ATK, MAFP Histone deacetylase Trichostatin, FK228 Phospholipase D Hexadecylphosphocholine Phosphatases Okadaic acid, fostreicin, calyculin A DNA synthesis Plk1 G2 Figure 11 Natural products and the cell cycle. Modified from L. Meijer, Le cycle de division cellulaire et sa régulation. Oncologie 2003, 5 (7–8), 311–326. Copyright Springer-Verlag 2003. Reprinted with permission. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 153 assay points per day in a number of cases), and the resultant screening capacity at many companies is significantly larger than the potential input from in-house chemical libraries. Since screening capacity is no longer the rate-limiting step, some major pharmaceutical companies became very interested in screening natural products (either as crude extracts or as prefractionated ‘peak libraries’) as a low-cost means of discovering novel lead compounds. A good illustration is the discovery at Merck Research Laboratories of a new antibiotic, through the testing of a library of 250 000 natural product extracts in a custom-designed assay involving an engineered strain of Staphylococcus aureus incorporating the fatty acid synthase pathway enzyme, FabF.137 Platensimycin is a selective FabF inhibitor and exhibits in vitro activity against several drug-resistant bacteria. Such promise has also spawned small companies such as Merlion Pharmaceuticals in Singapore, which has a library of many thousands of natural product extracts and a smaller number of pure natural products derived from a variety of sources, which it exposes to validated drug targets provided by pharmaceutical companies, with the goal of generating drug leads.138 Most of the screens used are proprietary and the published information is rare, although general summaries of this approach have been published.139 High-throughput assays are becoming less expensive, and such assays are moving from the industrial or industrial–academic consortium-based groups to academia in general, with specific expression systems being employed as targets for natural product lead discovery.140 The application of new techniques, including new fluorescent assays, NMR, affinity chromatography, and DNA microarrays, has led to significant advances in the effectiveness of high-throughput screening.141,142 3.06.4.1.2 Cell-based assays As mentioned in Section 3.06.4.1.1, the advent of new and robust high-throughput screens has had, and continues to have, a major impact on natural products research in the pharmaceutical industry. While some of the molecular target screens alluded to in Section 3.06.4.1.1 may involve use of transformed cells, the NCI’s 60 cell line cytotoxicity screen for antitumor agents represents a more traditional cell-based screen. It has been described in detail,143 and although this is not a true receptor-based screen, it has now been developed into a system whereby a large number of molecular targets within the cell lines may be identified by informatics techniques, and refinements are continuing. Information as to the current status of the screens involved can be obtained from the following URL: http://dtp.nci.nih.gov/. An assay based on differential susceptibility to genetically modified yeast strains has been described144 and has led to many screens based upon genetically modified yeasts, but at times, the low permeability of the unmodified yeast cell wall to chemical compounds has been overlooked. Thus, data from such screens, particularly those designed with gene deletions, must be carefully scrutinized since a large number are based upon hosts without a modified cell wall. In addition, there are simple but robust assays that can be used by workers in academia who do not have access to, or may not need, high-throughput screens. Examples are the brine shrimp and potato disc assays.145,146 3.06.4.1.3 In vivo assays Once the bioactive component has been obtained in pure form, either as a novel structure or as a known compound exhibiting previously unreported activity, then it must be assessed in a series of biological assays to determine its efficacy, potency, toxicity, and pharmacokinetics. These assays will help to determine the priority of the compound’s spectrum of activity within the portfolio of compounds that a group may be assessing as suitable for advanced development as either drug candidates or leads thereto. Knowledge of its putative mechanism of action (MOA) at this stage can also be a valuable discriminator in the prioritizing process. The NCI uses the Hollow Fiber Assay147 as a relatively inexpensive in vivo prescreen to prioritize compounds for testing in the more definitive human tumor xenograft models.148 Information on the in vivo assays currently used by the NCI, including a detailed description of the protocol used in the Hollow Fiber Assay, can be obtained from the following URL: http://dtp.nci.nih.gov/. 3.06.4.2 Tubulin Interactive Agents The majority of the tubulin interactive agents (TIAs) in development through to 2003, from preclinical studies up to clinical use, have been discussed in detail in a 2004 review by the authors,149 and also more recently (2005) in the book Anticancer Agents from Natural Products.150 The TIAs covered in that volume include taxanes 154 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 12 Tubulin interactive agents. (Taxol, 17; Figure 7),151 epothilones (Epo A, 32; Figure 10),152 and discodermolide (28; Figure 9),153 which act as promoters of polymerization of tubulin heterodimers to microtubules, leading to mitotic arrest through suppression of dynamic changes in microtubule functions. Other chapters are devoted to combretastatins (e.g., CA-4 phosphate, 39; Figure 12),154 vinca alkaloids (20, 21; Figure 7),155 maytansanoids (24; Figure 8),91 dolastatins (e.g., dolastatin 10, 31; Figure 9),117 halichondrins (29; Figure 9),116 and hemiasterlins (e.g., hemiasterlin A, 40; Figure 12),156 which act through inhibition of tubulin heterodimer polymerization. The coverage also includes agents derived from or synthetically modeled on those initial structures in order to develop drug candidates with improved solubilities, pharmacodynamics, or metabolic patterns, compared with the original natural products. Besides this review and the book chapters cited, the interested reader should consult relevant chapters in this series, together with the references given therein, for a discussion of the multiplicity of structures that have been developed from natural product lead compounds. Another detailed discussion of the marine-derived TIAs mentioned above (discodermolide, dolastatins, halichondrins, hemiasterlins) is presented in a review of natural products from marine invertebrates and microbes as modulators of antitumor targets.157 Other agents discussed in this review include dictyostatin, diazonamide (27; Figure 9), eleutherobin and laulimalide (41; Figure 12), which all act in a similar manner to the taxanes. While most TIAs act either as reversible inhibitors or promoters of tubulin heterodimer polymerization as mentioned above, pironetin (42; Figure 12), derived from a Streptomyces species, is the only TIA identified so far that acts through covalent binding to the -tubulin chain.158 The binding occurs at Lys352, an amino acid located at the entrance of a small pocket in -tubulin that faces the -tubulin of the next dimer.159 No derivatives have yet been reported as candidate leads. 3.06.4.3 Inhibitors of Topoisomerases I and II In early 2004 these authors reviewed new developments in the field of topoisomerase inhibitors in a special issue of the Journal of Natural Products honoring Drs. Monroe Wall and Mansukh Wani, the codiscoverers of both Taxol and camptothecin.160 The history of camptothecin (18; Figure 7) is presented in this review, and although the majority of new topoisomerase I inhibitors are based on the camptothecin pharmacophore,161 the Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 155 Figure 13 Topoisomerase and HDAC inhibitors. protein kinase inhibitor staurosporine (43; Figure 13) is also a topoisomerase inhibitor and various derivatives of the basic staurosporine scaffold inhibit both topoisomerases I and II.162 The anthracyclines are another class of important drugs that act via inhibition of topoisomerase II, with doxorubicin (44; Figure 13) being a prime example of the many members of this class.163 It should be pointed out, however, that almost all of the clinically useful compounds of this chemical class were developed as a result of their cytotoxic activities and without prior knowledge of this MOA.163 Likewise, the clinically active podophyllotoxin derivative, etoposide 156 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity (19; Figure 7), was developed by the then Sandoz company through modification of epi-podophyllotoxin without prior knowledge of the mechanism.164 It is interesting to note that podophyllotoxin acts as an inhibitor of tubulin polymerization, whereas etoposide acts on topoisomerase II. Although etoposide is a commonly used anticancer drug, acquired drug resistance and poor water solubility remain serious problems, and extensive research is being devoted to the production of a new generation of clinical trial candidates.164 A subsequent paper has reviewed the anticancer activity of some new topoisomerase inhibitors, which include 6 topoisomerase I, 12 topoisomerase II, and 6 dual topoisomerase inhibitors, many of which are derivatives of natural products.165 A second paper166 has reported on an analogue, AK-37 (45; Figure 13), of a marine-derived pyridoacridine, which stabilizes the topoisomerase I cleavable complex in a manner comparable to that of 9-nitro-camptothecin, which is currently in Phase III clinical trials for the treatment of pancreatic cancer in combination with gemcitabine. For those interested in reading further, the wide variety of structures and activities of pyridoacridines has been reviewed.167 3.06.4.4 Inhibitors of Histone Deacetylases The role of histone deacetylases (HDACs) in the regulation of gene expression, oncogenic transformation, cellular differentiation, and the promotion of angiogenesis is discussed by the authors157 and references cited therein. Suffice it to say, the inhibition of HDAC activity can exert a significant role in suppression of the neoplastic process. HDAC inhibitors have been described as tripartite: an enzyme-binding group, frequently aromatic; a hydrophobic spacer group; and an inhibitor group.168–170 Trichostatin A (TSA) (46; Figure 13) clearly demonstrates such a system, with the structure mimicking the Lys side chain of the substrate (the ‘linker’), the inhibitory end being the zinc-chelating hydroxamic acid, and the aromatic enzyme-binding group being the 4-dimethylaminobenzoyl group. This molecule, together with its congeners (trichostatins B, C, and D), was first isolated as an antifungal agent,171 and approximately a decade later they were found to have potent differentiation-inducing and antiproliferative activities in Friend erythroleukemia cells. Subsequently, TSA demonstrated potent in vitro and in vivo inhibition (nanomolar range) of class I and class II HDACs, with a slight selectivity for HDAC1 and HDAC6 compared to HDAC4. The S enantiomer of TSA was inactive, and neither enantiomer had any activity against the class III enzymes. The full MOA has not yet been elucidated, but a large series of effects were observed in signal transduction systems, including induction of apoptosis when healthy and tumor cells from many different sources were treated with this agent.172 Identification of the basic structural features of TSA and its initial activities led to research on the synthesis of compounds that were more stable and had improved water solubility. Early research with hexamethylene bisacetamide (HMBA) (47; Figure 13), belonging to a family of molecules known as hybrid polar compounds (HPCs), demonstrated that it induced hyperacetylation of histone H4 in healthy keratinocytes, as well as in squamous cell carcinoma derived from these cells, but did not inhibit their growth in vitro and induced a wide variety of other pathway modulations.173 The high doses of HMBA required for in vivo activity resulted in toxicity and led to cessation of development, but these results, combined with a knowledge of the basic structure of TSA, led to the development of a series of second-generation HPCs, which were tested as HDAC inhibitors. The lead compound from these studies, suberoylanilide hydroxamic acid (SAHA) (48; Figure 13), was approved in 2006 as vorinostat (Zolinza) and still is currently in over 40 clinical trials (phases I, II, and III), either as a single agent or in combination with other agents, against a variety of refractory tumors, both solid and leukemic in nature, including a Phase II study of an oral formulation.174 Efforts to resolve the problems of low yields of (R)-TSA from natural sources and difficulties in achieving its total synthesis have resulted a simple four-step strategy being devised for the synthesis of achiral amide analogues of the natural product. The analogues consisted of a hydroxamate function, a benzamide and an aliphatic spacer, with maximal inhibitory activity being observed with a five-carbon linker chain.175 The resulting lead compound was 6-(4-dimethylaminobenzoyl)-aminocaproic acid hydroxamide (49; Figure 13), and although the antitumor and cell transduction activities of these compounds have been reported, no in vivo data have yet been published.176 The natural product trapoxin (50; Figure 13) was reported to be an irreversible inhibitor of HDACs in 1993, but in contrast to TSA, it was found to demonstrate some selectivity against class I and class II HDACs, Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 157 inhibiting HDAC1 and HDAC4 but not HDAC6.177 Combination of structural features of trapoxin, TSA, and another potent HDAC inhibitor, the marine natural product psammaplin A (35; Figure 10), resulted in the de novo synthesis of NVP-LAQ-824 (dacinostat) (51; Figure 13), which inhibits HDAC and the proliferation of cancer cell lines at low nanomolar concentrations; it showed efficacy in a number of solid tumor xenograft models, advancing to Phase I clinical trials in 2002178,179 but was discontinued by Novartis in 2005. The full history of its evolution has been reviewed.180 The microbially derived depsipeptide, FR-901228 (romidepsin) (52; Figure 13), which was originally identified as a result of its potent antitumor activity, is now known to be active in signal transduction as a result of its HDAC activity,181 and it is currently in Phase I and Phase II clinical trials. 3.06.4.5 Protein Kinase Inhibitors Several agents that have advanced into clinical trials or commercial use in recent years have either been derived directly from nature or incorporate key structural features from natural products. Thus, the development of Gleevec can be traced back to ATP mimicry, with its history briefly reviewed by Newman et al.,182 and the history of Iressa is similar. 3.06.4.5.1 Flavopiridol The flavone, flavopiridol (Alvocidib) (53; Figure 14), is totally synthetic, but its novel structure is based on the natural product rohitukine (54; Figure 14), isolated from Dysoxylum binectariferum. It was originally considered to be an inhibitor of cyclin-dependent kinases (CDKs) (the regulators of the G2 to M transition in the cell cycle), and has entered into Phase I and then Phase II clinical trials against a broad range of tumors.183 Like the olomucine (55; Figure 14) derivative, roscovitine (selicicib) (56; Figure 14), it has now been reported to be a very potent inhibitor of CDK-7 and CDK-9, the kinases primarily responsible for promoting RNAP II (RNA polymerase II) activity, thus involving these agents in the transcription process. The molecular targets/ interactions involved in the transcription processes and flavopiridol interactions have been reviewed.184,185 Currently (early 2008), several single-agent Phase I and Phase II clinical trials against leukemias, lymphomas, and solid tumors are active, while over 10 Phase I and Phase II trials are active in combination with other anticancer agents. 3.06.4.5.2 Bryostatins The bryostatins are a class of highly oxygenated macrolides, and the multiyear program that culminated in the isolation and purification of (currently) 20 bryostatin structures, has been well documented by a number of authors over the years.186–193 These reviews may be consulted for the experimental details that indicated that the bryostatins, and in particular, bryostatin 1 (57; Figure 14), which has been the focus of preclinical and clinical studies, have signal transduction activities, and details on the clinical trials of bryostatin 1 are reviewed in Newman.193 While the total synthesis of bryostatin 1 is not a feasible process for the production of this agent, three of the naturally occurring bryostatins, bryostatins 7,194 2,195 and 3,196 have been synthesized, and their syntheses and the syntheses of other partial bryostatin structures, including bryostatin 1, have been reviewed;190,192,197 these reviews should be consulted for specific details of reaction schemes and comparisons of routes. None of these methods, however, are viable for the large-scale production of any of the bryostatins for further development. However, analytical studies by the Wender group of the potential binding site of the phorbol esters on protein kinase C (PKC) as a guide to the design of simpler analogue of these agents198 were expanded to bryostatin 1,199 and led to the production of simpler bryostatin analogues known colloquially as ‘bryologues’ that maintained the putative binding sites at the oxygen atoms at C1 (ketone), C19 (hydroxyl), and C26 (hydroxyl). These molecules (58, 59; Figure 14) demonstrated nanomolar binding constants when measured in displacement assays of tritiated phorbol esters, with the figures being in the same general range as bryostatin 1, and had activities in in vitro cell line assays close to those demonstrated by bryostatin 1 itself.200–203 Introduction of a second lactone gave a compound (60; Figure 14) with 8 nmol l1 binding affinity and an ED50 of 113 nmol l1 against P388,204 and the use of different fatty acid esters gave compounds exhibiting binding affinities for PKC isozymes in the 7–232 nmol l1 range depending upon the fatty acid used.205 A further simple modification 158 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity involving removal of a methyl group in the C26 side chain gave a compound (61; Figure 14) with the binding affinity to PKC at the picomolar level,206 and demonstrating greater potency than bryostatin 1 in in vitro cell line assays. Improved syntheses of the bryologues might well permit further exploration of these analogues.207,208 3.06.4.5.3 Adenine derivatives The observation that substituted purines, particularly 6-dimethylamino-purine (6-DMAP) (62; Figure 14) and isopentenyladenine (63; Figure 14), from Castanea species, showed weak inhibition of CDK1/cyclin B led to the search for other purine-derived compounds.209 Another plant secondary metabolite originally isolated from the cotyledons of the radish, and subsequently named olomucine (55; Figure 14), demonstrated an improved Figure 14 (Continued) Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 159 Figure 14 Protein kinase inhibitors efficacy (IC50 ¼ 7 mol l1) and selectivity for CDKs, and to some extent, MAP kinases, by direct competition with ATP. Olomucine, which earlier had been synthesized,210 disproved the existing dogma that no specific kinase inhibitors could be found for ATP-binding sites since they would be swamped by the presence of excess of ATP. Further development of this series using combinatorial chemistry techniques led to roscovitine (56; Figure 14), and finally to purvalanol A (64; Figure 14) and purvalanol B (65; Figure 14). The purvalanols demonstrated improved potency, with IC50 values in the range of 4–40 nmol l1, compared to 450 nmol l1 for roscovitine.211 The R-isomer of roscovitine is currently in Phase II clinical trials in Europe. Although some beneficial effects are observed with signal transduction inhibitors (STIs) alone, complete or partial responses tend only to be demonstrated when sequential treatments of STI/cytotoxin are used, so also with R-roscovitine, sequential treatment with cytotoxins is being used and/or considered. 3.06.4.5.4 Indigo and the indirubins Hydroxylation of indigo in the 3-position, presumably by a suitable cytochrome P-450, gives a product that is tautomeric with the 3-keto analogue, indoxyl (66; Figure 14), and various levels of oxidation then lead to a mixture of indigo, indirubin (67; Figure 14) and their isomers, which is commonly used as the source of indigo dyestuffs, a mixture obtained from the plant Isatis tinctoria found to contain an indigo precursor.212 Although usually considered to be plant products, indigo and the indirubins have been reported from four nominally independent sources: a variety of plants,212 a number of marine mollusks, usually belonging to the Muricidae family of gastropods,213 natural or recombinant bacteria,214 and human urine.215 The indirubins have been identified as the major active components of the TCM formulation known as Danggui Longhui Wan, which has been used for many years to treat chronic myelogenous leukemia (CML) in China.216 Of importance from both a natural product and a pharmacological perspective, the indirubins were recognized as being inhibitors of several CDKs and potent inhibitors of glycogen synthase kinase-3 (GSK-3).217 Included in this study were 6-bromoindirubin (68; Figure 14), first isolated from nature from the mollusk Hexaplex trunculus,209 and its chemically modified oxime derivative BIO (69; Figure 14), and these 160 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity two compounds demonstrated an at least fivefold specificity versus CDK1/cyclin B and/or CDK/p25, and significantly greater specificity against a wide range of other kinases. Significantly, GSK-3 is also an important target in both Alzheimer’s disease and type 2 diabetes, and although indole derivatives have not been reported as being associated with pharmacological intervention in these specific disease areas, their potential must be considered quite high. The treatment potential for inhibitors of GSK-3, including a listing of other natural product-related structures serving as possible inhibitors in these disease states, has recently been reviewed.218 Using the same basic suite of compounds, it was demonstrated that indirubins serve as ligands for the ‘orphan receptor’ known as the aryl hydrocarbon receptor (AhR).219 No other natural ligands have yet been identified for AhR, even though, contrary to earlier beliefs, it has existed for over 450 million years. Indole-containing compounds, however, had been suggested as natural ligands for AhR slightly earlier.220 Full details of the chemistry involved, and SARs established using X-ray crystallography and molecular modeling techniques, have been published.221 Among other natural products with indirubin-like kinase inhibitory activities are the meridianins (e.g., meridianin A) (70; Figure 14), a group of halogenated indole derivatives that are closely related to the base structures of variolin B (7; Figure 4), the psammopemmins (e.g., psammopemmin A) (71; Figure 14) and discodermindol (72; Figure 14). Variolin B, the psammopemmins and discodermindol were isolated from sponges, whereas the meridianins were isolated from the ascidian Aplidium meridianum.222 3.06.4.6 Protein Folds and Inhibitors of Kinases and Phosphatases Significant effort has been, and continues to be, devoted to the development of novel kinase inhibitors through the ‘‘fitting of structures to the ATP-binding sites,’’ and this approach has been quite successful in producing structures for clinical trials.223 However, the Waldman group has successfully developed a variation on this theme in which, rather than initially concentrating on the specifics of the ATP-binding site, they have used two other fundamental premises to search for kinase, and other enzyme inhibitors. First, they considered that biologically active natural products are viable, biologically validated starting points for library design, permitting the discovery of lead compounds possessing an enhanced probability of success if included in high-throughput screening;125,127 and second, that although estimates of the number proteins in humans range between 100 000 and 450 000, the number of topologically distinct shapes, defined as protein folds, is actually much lower, with estimates of 600–8000.224 Thus, if an inhibitor of a specific protein fold from nature could be found, then it could be used as a prototype for the development of closely related structures that may inhibit proteins with similar folds, and even allow for the discovery of specificity. These concepts are fundamentally similar to the privileged structure concept mentioned in Section 3.06.3.5.3, but the Waldmann approach has the added dimension of using protein folding patterns as the basis for subsequent screens. The success of this approach was demonstrated by the derivation of inhibitors of Tie-2, insulin-like growth factor 1 receptor (IGF-1R), and vascular endothelial growth factor receptors 2 and 3 (VEGFR-2 and VEGFR-3), from the original discovery of the Her-2/Neu inhibitor, nakijiquinone C (73; Figure 15). Derived from a marine sponge and first reported by Kobayashi et al.225 in 1995, nakijiquinone C was shown to be an inhibitor of epidermal growth factor receptor (EGFR), c-ErbB2, and PKC, in addition to having cytotoxic activity against L1210 and KB cell lines. Testing of a library of 74 compounds, built around the basic nakijiquinone C structure, against a battery of kinases with similar protein domain folds, yielded seven new inhibitors with low micromolar activity in vitro, including one VEGFR-2 inhibitor (74; Figure 15) and four inhibitors of Tie-2 kinase (75–78; Figure 15), a protein intimately involved in angiogenesis, and for which, at the beginning of the study, no inhibitors were known.124 During the study, the first natural product inhibitor of Tie-2 kinase was reported226 (79; Figure 15) from the plant Acacia aulacocarpa, and a set of four papers from another research group demonstrated the activity of synthetic pyrrolo[2,3-d]pyrimidines as inhibitors of the same class of kinases.227–230 The details of the models used, the chemistry leading to the nakijiquinone-based compounds, and the ribbon structures of the kinase domain of the insulin receptor, with the corresponding homology domains of the as yet uncrystallized VEGFR-2 and Tie-2, have been fully reviewed.129,231 A similar approach has been used in the identification of phosphatase inhibitors. Postulating that the -hydroxy-butenolide group of the marine-derived metabolite, dysidiolide (33; Figure 10), was the major determinant of phosphatase activity, testing of a 147-member library built around this molecule yielded a Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 15 Kinase, phosphatase, and Hsp90 inhibitors. 161 162 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity compound (80; Figure 15) 10-fold more potent (IC50 ¼ 350 nmol l1) than the parent against Cdc25A.125 In addition, other members of the library were identified with low micromolar activities against the enzymes, acetylcholinesterase and 11 -hydroxysteroid dehydrogenase type 1, which fall within the same ‘similarity cluster’ as Cdc25A.232 3.06.4.7 Inhibitors of Heat Shock Protein 90 Heat shock protein 90 (Hsp90) is a chaperone protein that plays an important role in stabilizing the conformation of many cell-signaling proteins and maintaining their function. In this respect, many oncogenic proteins are more dependent on Hsp90 than their normal counterparts, and hence Hsp90 plays an important role in maintaining transformation and increasing the survival and growth tendency of cancer cells. It has also been shown to exist in an activated form in cancer cells while existing in a latent inactive form in normal cells, thus making it an attractive target for chemotherapy in cancer and other diseases, such as neurodegenerative diseases.233 Advances in the development of Hsp90 inhibitors have been reviewed.234,235 3.06.4.7.1 Ansamycins: geldanamycin derivatives The development of the ansamycins leading to the 17-substituted analogues has been reviewed.236 This review highlights the significant differences in the macrocyclic ring stereochemistries reported in the literature for what is nominally the same molecule. These differences are not simply due to a complete stereochemical inversion around the ring, where the relative stereochemistries are maintained, but are quite different renditions from different research groups and should be noted when referring to different papers. 17-Allylaminogeldanamycin (17-AAG; tanespimycin) (81; Figure 15) is in over 10 phases I, II, and III clinical trials against leukemias, lymphomas, and solid tumors, either as a single agent or in combination with other agents. The more soluble material, 17-dimethylamino-ethylaminogeldanamycin (17-DMAG) (82; Figure 15), is in Phase I clinical trials as a single agent against solid tumors, while a number of other 17-substituted derivatives have been prepared as potential alternative candidates.237,238 Two apparent anomalies in the interactions of geldanamycin (GA) derivatives and radicicol (monorden) (83; Figure 15) with Hsp90 have been under intensive study. The first anomaly is that, despite the fact that both healthy and tumor cells require Hsp90 for cellular function, they respond differently to these drugs, and the second is the fact that the affinity of these drugs for recombinant Hsp90 (rHsp90) is much lower than the levels required for responses in tumor cell lysates. The higher binding affinity for Hsp90 in tumor lysates has been attributed to the existence of other cochaperones in tumor cells that are not expressed in healthy cells, and this effect was demonstrated by the addition of such proteins to rHsp90.239 In addition, X-ray crystal studies have demonstrated that the structure of GA in the unbound form has a trans-configuration at the amide bond between the benzoquinone and the rest of the ansa ring, whereas when bound to Hsp90, GA displays the cis-configuration at this center.240 Similarly, Jez et al.241 reported that the closely related GA derivative 17-DMAG requires both a macrocyclic ring conformational change and a trans–cis isomerization of the amide bond in order to bind to Hsp90. The tumor selectivity, however, is still a subject of investigation.242 3.06.4.7.2 Non-ansamycin inhibitors Supply problems associated with GA derivatives and radicicol, together with GA toxicity problems, led Chiosis et al. to propose the use of a simple substituted adenine derivative as a potential base molecule. Significantly, the proposal was based on considering which particular substructures might provide ATP mimics with improved binding characteristics, rather than computerized modeling. Thus, knowledge of the requirements of the ATP-binding pocket of Hsp90, and demonstration that a small molecule could function as a cytostatic agent,243 provided the intellectual stimulus for designing the purine-based PU class of compounds.244–246 Rational changes in the substituents in both rings and alteration of the length and rigidity of the linker gave rise to PU24FCl (84; Figure 15),247 which, although not the most active in the series, was utilized to further investigate Hsp90 inhibition in both healthy and tumor cells. The extensive effects exhibited by both healthy and tumor tissues when exposed to the compound have been reported,248 and, as with 17-AAG and GA, PU24FCl exhibited at least 10- (brain, pancreas, lung) to 50-fold (heart, kidney, liver) lower affinity for Hsp90s from healthy tissues as compared to those from transformed cells. Later studies have shown that replacement of Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 163 the methylene bridge with sulfur gives 8-arylsulfanyl adenine derivatives (e.g., 85; Figure 15) of greater potency,249 while introduction of an ionizable amino group in the N(9) side chain improved both the water solubility and potency of the compounds to give orally active agents (e.g., 86; Figure 15).250,251 3.06.4.8 Proteasome Inhibitors The proteasome is a multienzyme complex involved in the ubiquitin–proteasome pathway control of cell-cycle progression, in the termination of signal transduction cascades, and in the removal of mutant, damaged, and misfolded proteins. As such, it is a promising therapeutic target, and the background to this aspect has been reviewed.252–255 The synthetic dipeptidyl peptide boronate, bortezomib (Velcade) (87; Figure 16), is the first clinical drug that uses this MOA,256–258 and the development of this compound, which is based upon a natural product-derived structure that inhibited chymotrypsin, has been described by the original inventor.259 There are however a significant number of other compounds from nature, and their derivatives, that have led to a greater understanding of the intricacies of this multienzyme complex. The 20S proteasome in mammals has three closely linked proteolytic activities, which are termed trypsin-, chymotrypsin-, and caspase-like from their substrate profiles, although the complex only acts as a concerted whole; individual activities are not demonstrable. In fact, if the chymotrypsin-like activity is inhibited by a suitable compound then a large reduction in the rate of protein degradation is observed, but if the sites corresponding to the other nominal activities are modified, the overall rate of hydrolysis of proteins is not significantly changed. Owing to the substrate specificity of chymotryptic sites, most inhibitors are hydrophobic, whereas in the case of the other two active sites, their ‘peptide-based’ substrates/inhibitors tend to be charged. As a result, almost all of the proteasome inhibitors tend to have chymotrypsin-like activities with some overlapping, but weaker, effects on the other sites. In 1991, the microbial metabolite lactacystin (88; Figure 16) was reported to induce neuritogenesis in neuroblastoma cells,260 and this was followed by reports261,262 demonstrating that radio-labeled lactacystin selectively modified the 5(X) subunit of the mammalian proteasome, and irreversibly blocked activity. In subsequent studies, it was demonstrated263,264 that the actual inhibitor in vitro was the -lactone, clastolactacystin- -lactone (89; Figure 16), and that this substance was formed spontaneously when lactacystin was exposed to neutral aqueous media. The parent compound and other analogues have been synthesized, and the authors suggested that clasto-lactacystin- -lactone should be named omuralide (89; Figure 16).265,266 The marine bacterial metabolite salinosporamide A (Section 3.06.3.3.5) (22; Figure 8) demonstrates activity as a cytotoxic proteasome inhibitor86 and has been synthesized.267 Compared to omuralide, salinosporamide is uniquely functionalized and has a cyclohexene ring replacing the isopropyl group found at the C(5)-position in omuralide. The isopropyl group in omuralide is essential for the activity, so salinosporamide A might interact with the 20S proteasome in a modified manner. This molecule is being developed by Nereus Pharmaceuticals, and currently is in Phase I clinical trials against refractory lymphomas and myelomas, as well as various solid tumors. The epoxyketone microbial metabolites epoxomicin (90; Figure 16) and eponemycin (91; Figure 16) exhibited cytotoxic activities as a result of proteasome inhibition,268,269 being the most selective proteasome inhibitors reported to date. There are reports of other natural products active as proteasome inhibitors but with different mechanisms to those above. Thus, the cyclic peptide TMC-95-A (92; Figure 16), isolated from Apiospora montagnei, is a potent chymotrypsin-like inhibitor, but with activity against the other sites as well,270 apparently binding noncovalently to active sites through an array of hydrogen bonds. ()-Epigallocatechin 3-gallate (93; Figure 16) is a potent covalent inhibitor of the 20S proteasome, apparently due to acylation of the active site threonines through threonine cleavage of the ester linkage in EGCG.271 3.06.4.9 DNA Interactive Agents (Non-Topoisomerases I and II Inhibitors) The complex alkaloid ecteinascidin 743 (Et-743, Yondelis) (94; Figure 16), discovered from the colonial tunicate Ecteinascidia turbinata,272,273 was found to have a unique MOA, binding to the minor groove of DNA and interfering with cell division, the genetic transcription processes, and DNA repair machinery.274,275 There has been a considerable number of reports published in the literature giving possibilities as to the MOA(s) of 164 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Figure 16 Proteasome inhibitors and DNA interactive agents. Et-743 when tumor cells are treated in vitro. A significant problem with some of the reports is that the concentration(s) used in the experiments are often orders of magnitude greater than those that demonstrate activity in vivo. These levels are in the low nanomolar to high picomolar range and thus care should be taken when evaluating published work on the MOA of this compound. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 165 The issue of compound supply for advanced studies was solved by the development of a semisynthetic route from the microbial product cyanosafracin B (95; Figure 16),276 and this and other aspects of the discovery and development have been comprehensively reviewed.157,277 Under the name Yondelis, Et-743 has been granted orphan drug designation in Europe and the United States, and was approved by the European Medicines Agency (EMEA) in late September 2007 for the treatment of soft tissue sarcomas (STS).278 It is also in Phase II and Phase III trials in ovarian, metastatic breast and prostate cancers, and pediatric sarcomas. 3.06.4.10 Caspase Activation and Apoptosis Induction The relatively simple naphthoquinone -lapachol (96; Figure 17) is a well-known compound obtained from the bark of the lapacho tree, Tabebuia avellanedae, and other species of the same genus that are native to South America. -Lapachol and other plant components are extensively used as ethnobotanical treatments in the Figure 17 Inhibitors of caspase activation, apoptosis induction, HIF, and miscellaneous targets. 166 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Amazonian region, and -lapachol was advanced to clinical status by the NCI in the 1970s. It was later withdrawn due to unacceptable levels of toxicity, but its close relative -lapachone (97; Figure 17) has demonstrated interesting molecular target activity, with one MOA being the induction of apoptosis in transformed cells.279 Evidence of its involvement in transcription processes has been reported demonstrating that the agent induced activation of caspase-3, inhibition of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-B) and subsequent downregulation of bcl-2.280 Currently, -lapachone (ARQ501) is in Phase II clinical trials in the United States for advanced solid tumors, and further information on the background of these agents may be obtained from the 2004 review.281 3.06.4.11 Hypoxia Inducible Factor Hypoxia inducible factor 1 (HIF-1) is composed of two subunits, an oxygen-sensitive inducible factor (HIF-1 ) and the constitutive HIF-1 (also known as AhR nuclear translocator (ARNT)), which may prove to be an important target in diseases that have a hypoxic component such as cancer (where the interior of a tumor is anoxic compared with the outer surfaces), heart disease, and/or stroke. The involvement of HIF proteins with a variety of inhibitors (not necessarily direct inhibition, but alteration of transduction pathways upstream and downstream) have been reviewed, and included in the review are well-known materials with natural product ‘backgrounds’, such as Taxol., vincristine, 2-methoxyestradiol, rapamycin, GA, quinocarmycin, and the IP3K inhibitors, wortmannin and LY-294002.282 Of significance from a natural product perspective was the initial realization that inhibition of thioredoxin reductase 1 (TRX-1) may act indirectly on HIF-1 . By comparing the NCI 60 human cancer cell line cytotoxicity profile of a known TRX-1 inhibitor and Phase II clinical candidate, PX-12 (98; Figure 17), with the profiles of a range of compounds in the NCI screening database, the fungal natural product pleurotin (99; Figure 17) was identified as exhibiting a similar killing pattern to PX-12.283 Research on a focused combinatorial library of naphthoquinone acetals based upon palmarumycin CP1 (100; Figure 17), which included diepoxins (e.g., diepoxin, 101; Figure 17) and deoxypreussomerins (e.g., deoxypreussomerin A, 102; Figure 17), indicated that they possessed potent cytotoxicity, but their potential targets were unidentified at that time.284 Palmarumycin CP1, however, was later shown to have inhibitory activity comparable to that of pleurotin in the TRX-1 assay, with IC50 values in the range of 170–350 nmol l1, and it was demonstrated that certain aspects of the base structure, in particular the enone system, were required for activity in this assay.285 Evidence for direct inhibition of HIF-1 by both pleurotin and PX-12, helped to demonstrate that the cytotoxicity of these compounds, and hence palmarumycin CP1, was likely due to HIF-1 interactions.286 Further palmarumycins isolated from extracts of the fermentation broth of an unidentified ascomycete from Costa Rica failed to show activity in the assays used, but provided important SAR information.287 This information in turn led to further modifications of the base structure, yielding the simple analogues S-11 (103; Figure 17) and S-12 (104; Figure 17), which exhibited biological activities comparable to pleurotin in both the thioredoxin enzyme (TRX-1) system and (most importantly) in the cytotoxicity assays.287 Thus, a fairly complex interaction of results from several different research groups has led to promising candidates for further biological studies, including in vivo experiments that are planned and will be reported in due course. 3.06.4.12 Miscellaneous Target Inhibitors There are a number of agents particularly from marine sources, whose initial molecular targets have been identified although it is highly probable that over the next few years, these initial targets will be refined as methods and other information becomes available. One such compound, aplidine, is an agent with multiple targets. Formally, dehydrodidemnin B (105; Figure 17), it was first reported in a patent and then referred to in a 1996 paper on SARs among the didemnins.288 In 1996, the antitumor potential was reported by PharmaMar scientists and the total synthesis was reported in a patent application in 2000 and the patent was issued in 2002. The compound was advanced to Phase I clinical trials in 1999 under the trade name of Aplidin for the treatment of both solid tumors and non-Hodgkin’s lymphoma and published details through early 2004 are given in Newman and Cragg28 together Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 167 with discussion as to the mechanisms of action that might be relevant. Details of the progress of this drug through preclinical and clinical development have been reviewed.28,277,289 It should be noted that the clinical trials of the very close aplidine analogue didemnin B (106; Figure 17) were discontinued because of the toxicities observed, including significant immunosuppression. In contrast, evidence for the lack of myelosuppression by aplidine was reported using a murine competitive repopulating model as the test system,290 and no hematological toxicity has been observed clinically.277 It is very interesting both chemically and pharmacologically that the removal of two hydrogen atoms, that is, conversion of the lactyl side chain to a pyruvyl side chain, appears to significantly alter the toxicity profile, as this is the only formal change in the molecule when compared to didemnin B. However, the comments on dosage regimens should be taken into account when such comparisons are made in the future.291 3.06.5 Summary and Future Prospects Nature has been a source of medicinal products for millennia, and during the past century many useful drugs have been developed from natural sources, particularly plants. It is clear that nature will continue to be a major source of new drug leads. The drug potential of the marine environment remains relatively unexplored, but it is becoming increasingly evident that the realm of microorganisms offers a vast untapped potential. With the advent of genetic techniques that permit the isolation and expression of biosynthetic cassettes, microbes and their marine invertebrate hosts may well be the new frontier for natural products lead discovery. Plant endophytes also offer an exciting new resource, and research continues to reveal that many of the important drugs originally thought to be produced by plants are actually products of endophytic microbes residing in the tissues between living plant cells. This has been further accentuated by the recent report of the isolation of hypericin from an endophytic fungus from Hypericum perforatum.292 Effective drug development will depend on multidisciplinary collaboration embracing natural product lead discovery and optimization through the application of total and DOS and combinatorial chemistry and biochemistry, combined with good biology. The impressive number of anticancer drugs that are derived from natural sources are discussed in terms of their mechanisms of action, and as can be seen from these discussions, natural products from all sources still have the potential to lead chemists of all types into areas of drug discovery and development that would never have been considered if the ‘privileged structures from nature’ had not been isolated, purified, and used as probes of cellular and molecular mechanisms. In spite of the discussions in the early to late 1990s concerning the vast potential of combinatorial chemistry as a discovery tool, it is now quite evident that this technique, except in the very special cases of peptides and nucleosides (which are actually ‘privileged structures’ in their own right), is not the panacea that it was thought to be. However, the application of combinatorial synthetic methodology as a means to elaborate around a skeleton from a privileged structure demonstrates that the use of both techniques will lead to novel agents having potential as drug entities in many disease states.293 References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. J. K. Borchardt, Drug News Perspect. 2002, 15, 187–192. K. C. Huang, The Pharmacolog of Chinese Herbs, 2nd ed.; CRC Press: Boca Raton, FL, 1999. L. D. Kapoor, CRC Handbook of Ayurvedic Medicinal Plants; CRC Press: Boca Raton, FL, 1990. S. Dev, Environ. Health Persp. 1999, 107, 783–789. T. Johnson, CRC Ethnobotany Desk Reference; CRC Press: Boca Raton, FL, 1999. D. E. Moerman, Medicinal Plants of Native America; University of Michigan Museum of Anthropology: Ann Arbor, MI, 1986; Vol. 1. N. R. Farnsworth; R. O. Akerele; A. S. Bingel; D. D. Soejarto; Z. Guo, Bull. World Health Organ. 1985, 63, 965–981. D. S. Fabricant; N. R. Farnsworth, Environ. Health Perspect. 2001, 109 (Suppl.), 69–75. A. D. Buss; R. D. Waigh, Natural Products as Leads for New Pharmaceuticals. In Burger’s Medicinal Chemistry and Drug Discovery. Principles and Practice, 5th ed.; M. E. Wolff, Ed.; John Wiley & Sons, Inc.: New York, 1995; Vol. 1, pp 983–1033. C. Wongsrichanalai; A. L. Pickard; W. H. Wernsdorfer; S. R. Meshnick, Lancet Infect. Dis. 2002, 2, 209–218. D. L. Klayman, Science 1985, 228, 1049–1055. P. M. O’Neill; G. H. Posner, J. Med. Chem. 2004, 47, 2945–2964. 168 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 13. J. L. Vennerstrom; S. Arbe-Barnes; R. Brun; S. A. Charman; F. C. K. Chiu; J. Chollet; Y. Dong; A. Dorn; D. Hunziker; H. Matile; K. McIntosh; M. Padmanilayam; J. Santo Tomas; C. Scheurer; B. Scorneaux; Y. Tang; H. Urwyler; W. Sergio; W. N. Charman, Nature 2004, 430, 900–904. 14. G. H. Posner; I.-H. Paik; W. Chang; K. Borstnik; S. Sinishtaq; A. S. Rosenthal; T. A. Shapiro, J. Med. Chem. 2007, 50, 2516–2519. 15. J. L. Hartwell, Plants Used Against Cancer; Quarterman: Lawrence, MA, 1982. 16. G. M. Cragg; M. R. Boyd; J. H. Cardellina, II; D. J. Newman; K. M. Snader; T. G. McCloud, Ethnobotany and Drug Discovery: The Experience of the US National Cancer Institute. In Ethnobotany and the Search for New Drugs; D. J. Chadwick, J. Marsh, Eds.; Ciba Foundation Symposium, John Wiley & Sons Inc.: New York, 1994; Vol. 185, pp 178–196. 17. D. J. Newman; G. M. Cragg, J. Nat. Prod. 2007, 70, 461–477. 18. F. C. Schröder; J. J. Farmer; A. B. Attygalle; S. R. Smedley; T. Eisner; J. Meinwald, Science 1998, 281, 428–431. 19. T. L. Czaran; R. F. Hoekstra; L. Pagie, Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 786–790. 20. A. Fitter, Science 2003, 301, 1337–1338. 21. H. P. Bais; R. Vepachedu; S. Gilroy; R. M. Callaway; J. M. Vivanco, Science 2003, 301, 1377–1380. 22. S. A. Rice; D. McDougald; N. Kumar; S. Kjelleberg, Curr. Opin. Investig. Drugs 2005, 6, 178–184. 23. S. Borman, Chem. Eng. News 2005, 83 (5), 38. 24. G. Bulaj; O. Buczek; I. Goodsell; E. C. Jiminez; J. Kranski; J. S. Nielsen; J. E. Garrett; B. M. Olivera, Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 14562–14568. 25. M. S. Wallace, Expert Rev. Neurother. 2006, 6, 1423–1428. 26. M. F. Balandrin; A. D. Kinghorn; N. R. Farnsworth, Plant-Derived Natural Products in Drug Discovery and Development: An Overview. In Human Medicinal Agents from Plants; A. D. Kinghorn, M. F. Balandrin, Eds.; American Chemical Society: Washington, DC, 1993; Vol. 534, pp 2–12. 27. I. Raskin; D. M. Ribnicky; S. Komarnytsky; N. Ilic; A. Poulev; N. Borisjuk; A. Brinker; D. A. Moreno; C. Ripoll; N. Yakoby; J. M. O’Neal; T. Cornwell; I. Pastor; B. Fridlander, Trends Biotechnol. 2002, 20, 522–531. 28. D. J. Newman; G. M. Cragg, J. Nat. Prod. 2004, 67, 1216–1238. 29. D. J. Newman; R. T. Hill, J. Ind. Microbiol. Biotechnol. 2006, 33, 539–544. 30. N. B. Perry; L. Ettouati; M. Litaudon; J. W. Blunt; M. H. G. Munro, Tetrahedron 1994, 50, 3987–3992. 31. G. Trimurtulu; D. J. Faulkner; N. B. Perry; L. Ettouati; M. Litaudon; J. W. Blunt; M. H. G. Munro; G. B. Jameson, Tetrahedron 1994, 50, 3993–4000. 32. A. Ahaidar; D. Fernández; G. Danelón; C. Cuevas; I. Manzanares; F. Albericio; J. A. Joule; M. Álvarez, J. Org. Chem. 2003, 68, 10020–10029. 33. T. Diyabalanage; C. D. Amsler; J. B. McClintock; B. J. Baker, J. Am. Chem. Soc. 2006, 128, 5630–5631. 34. K. C. Nicolaou; R. Guduru; Y.-P. Sun; B. Banerji; D. Y. K. Chen, Angew. Chem. Int. Ed. 2007, 46, 5896–5900. 35. A. Poulev; J. M. O’Neal; S. Logendra; R. B. Pouleva; V. Timeva; A. S. Garvey; D. Gleba; I. S. Jenkins; B. T. Halpern; R. Kneer; G. M. Cragg; I. Raskin, J. Med. Chem. 2003, 49, 2542–2547. 36. E. McCoy; S. E. O’Connor, J. Am. Chem. Soc. 2006, 128, 14276–14277. 37. N. R. Pace, Science 1997, 276, 734–740. 38. M. T. Madigan; J. M. Martinko; J. B. Parker, Biology of Microorganisms, 8th ed.; Prentice-Hall: Upper Saddle River, NJ, 1996. 39. J. B. McAlpine; B. O. Bachmann; M. Piraee; S. Tremblay; A.-M. Alarco; E. Zazopoulos; C. M. Farnet, J. Nat. Prod. 2005, 68, 493–496. 40. S. Lautru; R. J. Deeth; L. Bailey; G. M. Challis, Nat. Chem. Biol. 2005, 1, 265–269. 41. K. Zengler; G. Toledo; M. Rappe; J. Elkins; E. J. Mathur; J. M. Short; M. Keller, Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15681–15686. 42. D. W. Udwary; L. Zeigler; R. N. Asolkar; V. Singan; A. Lapidus; W. Fenical; P. R. Jensen; B. S. Moore, Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 10376–10381. 43. E. A. Gontang; W. Fenical; P. R. Jensen, Appl. Environ. Microbiol. 2007, 73, 3272–3282. 44. M. R. Rondon; P. R. August; A. D. Bettermann; S. F. Brady; T. H. Grossman; M. R. Liles; K. A. Loiacono; B. A. Lynch; I. A. MacNeil; C. Minor; C. L. Tiong; M. Gilman; M. S. Osburne; J. Clardy; J. Handelsman; R. M. Goodman, M., G. R. App. Environ. Microbiol. 2000, 66, 2541–2547. 45. J. C. Venter; K. Remington; J. F. Heidelberg; A. L. Halpern; D. Rusch; J. A. Eisen; D. Wu; I. Paulsen; K. E. Nelson; W. Nelson; D. E. Fouts; S. Levy; A. H. Knap; M. W. Lomas; K. Nealson; O. White; J. Peterson; J. Hoffman; R. Parsons; H. Baden-Tillson; C. Pfannkoch; Y.-H. Rogers; H. O. Smith, Science 2004, 304, 66–74. 46. S. Yooseph; G. Sutton; D. B. Rusch; A. L. Halpern; S. J. Williamson; K. Remington; J. A. Eisen; K. B. Heidelberg; G. Manning; W. Li; L. Jaroszewski; P. Cieplak; C. S. Miller; H. Li; S. T. Mashiyama; M. P. Joachimiak; C. van Belle; J.-M. Chandonia; D. A. Soergel; Y. Zhai; K. Natarajan; S. Lee; B. J. Raphael; B. Bafna; R. Friedman; S. E. Brenner; A. Godzik; D. Eisenberg; J. E. Dixon; S. S. Taylor; R. L. Strausberg; M. Frazier; J. Craig Venter, PLoS Biol. 2007, 5 (3), e16. doi:10.1371/ journal.pbio.0050016. Published 13 March 2007. 47. F. Warnecke; P. Luginbühl; N. Ivanova; M. Ghassemian; T. H. Richardson; J. T. Stege; M. Cayouette; A. C. McHardy; G. Djordjevic; N. Aboushadi; R. Sorek; S. G. Tringe; M. Podar; H. G. Martin; V. Kunin; D. Dalevi; J. Madejska; E. Kirton; D. Platt; E. Szeto; A. Salamov; K. Barry; N. Mikhailova; N. C. Kyrpides; E. G. Matson; E. A. Ottesen; X. Zhang; M. Hernández; C. Murillo; L. G. Acosta; I. Rigoutsos; G. Tamayo; B. D. Green; C. Chang; E. M. Rubin; E. J. Mathur; D. E. Robertson; P. Hugenholtz; J. R. Leadbetter, Nature 2007, 450, 560–565. 48. L. Fieseler; U. Hentschel; L. Grozdanov; A. Schirmer; G. Wen; M. Platzer; S. Hrvatin; D. Butzke; K. Zimmermann; J. Piel, Appl. Environ. Microbiol. 2007, 73, 2144–2155. 49. C. Holden, Science 2005, 307, 1558. 50. H. B. Bode; R. Muller, Angew. Chem. Int. Ed. 2005, 44, 6828–6846. 51. A. H. Banskota; J. B. Mcalpine; D. Sørensen; A. Ibrahim; M. Aouidate; M. Piraee; A. M. Alarco; C. M. Farnet; E. Zazopoulos, J. Antibiot. 2006, 59, 533–542. 52. K. S. Lam, Trends Microbiol. 2007, 15, 279–289. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 53. 54. 55. 56. 57. 58. 59. 60. 61. 62. 63. 64. 65. 66. 67. 68. 69. 70. 71. 72. 73. 74. 75. 76. 77. 78. 79. 80. 81. 82. 83. 84. 85. 86. 87. 88. 89. 90. 91. 92. 93. 94. 95. 96. 97. 98. 99. 100. 101. 102. 103. 104. 105. 106. 107. 169 J. Clardy; M. A. Fischbach; C. T. Walsh, Nat. Biotech. 2006, 24, 1541–1550. J. W. Bok; D. Hoffmeister; L. A. Maggio-Hall; R. Murillo; J. D. Glasner; N. P. Keller, Chem. Biol. 2006, 13, 31–37. D. Hoffmeister; N. P. Keller, Nat. Prod. Rep. 2007, 24, 393–416. S. Rachid; K. Gerth; I. Kochems; R. Muller, Mol. Microbiol. 2007, 63, 1783–1796. F. Abe; K. Horikoshi, Trends Biotechnol. 2001, 19, 102–108. A. Amato, Chem. Eng. News 2006, 84, 14. A. Persidis, Nature Biotechnol. 1998, 16, 593–594. M. Rossi; M. Ciaramella; R. Cannio; F. M. Pisani; M. Moracci; S. Bartolucci, J. Bacteriol. 2003, 185, 3683–3689. P. L. Short, Chem. Eng. News 2007, 85(4), 20–21. R. Cavicchioli; K. S. Siddiqui; D. Andrews; K. R. Sowers, Curr. Opin. Biotechnol. 2002, 13, 253–261. J. Gomes; W. Steiner, Food Technol. Biotechnol. 2004, 42. A. Hoyoux; V. Blaise; T. Collins; S. D’Amico; E. Gratia; A. L. Huston; J. C. Marx; G. Sonan; Y. X. Zeng; G. Feller; C. Gerday, J. Biosci. Bioeng. 2004, 98, 317–330. C. Schiraldi; M. De Rosa, Trends Biotechnol. 2002, 20, 515–521. B. van den Burg, Curr. Opin. Microbiol. 2003, 6, 213–218. J. Wiegel; V. V. Kevbrin, Biochem. Soc. Trans. 2004, 32, 193–198. D. B. Johnson; K. B. Hallberg, Res. Microbiol. 2003, 154, 466–473. A. A. Stierle; D. B. Stierle; K. Kemp, J. Nat. Prod. 2004, 67, 1392–1395. D. B. Stierle; A. A. Stierle; D. Hobbs; J. Stokken; J. Clardy, Org. Lett. 2004, 6, 1049–1052. A. A. L. Gunatilaka, J. Nat. Prod. 2006, 69, 509–526. G. Strobel; B. Daisy; U. Castillo; J. Harper, J. Nat. Prod. 2004, 67, 257–268. R. X. Tan; W. X. Zou, Nat. Prod. Rep. 2001, 18, 448–459. D. Ezra; U. F. Castillo; G. A. Strobel; W. M. Hess; H. Porter; J. B. Jensen; M. A. Condron; D. B. Teplow; J. Sears; M. Maranta; M. Hunter; B. Weber; D. Yaver, Microbiology 2004, 150, 785–793. G. X. Zhou; E. M. K. Wijeratne; D. Bigelow; L. S. Pierson, III; H. D. VanEtten; A. A. L. Gunatilaka; Aspochalasins I, J, and K, J. Nat. Prod. 2004, 67, 328–332. A. Stierle; G. Strobel; D. Stierle, Science 1993, 260, 214–216. J.-Y. Li; R. S. Sidhu; A. Bollon; G. A. Strobel, Mycolog. Res. 1998, 102, 461–464. S. C. Puri; V. Verma; T. Amna; G. N. Qazi; M. Spiteller, J. Nat. Prod. 2005, 68, 1717–1719. T. Amna; S. C. Puri; V. Verma; J. P. Sharma; R. K. Khajuria; J. Musarrat; M. Spiteller; G. N. Qazi, Can. J. Microbiol. 2006, 52, 189–196. A. L. Eyberger; R. Dondapati; J. R. Porter, J. Nat. Prod. 2006, 69, 1121–1124. S. C. Puri; A. Nazir; R. Chawla; R. Arora; S. Riyaz-ul-Hasan; T. Amna; B. Ahmed; V. Verma; S. Singh; R. Sagar; A. Sharma; R. Kumar; R. K. Sharma; G. N. Qazi, J. Biotech. 2006, 122, 494–510. B. Guo; H. Li; L. Zhang, J. Yunnan Univ. 1998, 20, 214–215. L. Q. Zhang; B. Guo; H. Li; S. Zeng; H. Shao; S. Gu; R. Wei, Zhong Cao Yao (Chinese Tradit. Herb. Drugs) 2000, 31, 805–807. X. Yang; L. Zhang; B. Guo; S. Guo, Zhong Cao Yao (Chinese Tradit. Herb. Drugs) 2004, 35, 79–81. T. J. Mincer; P. R. Jensen; C. A. Kauffman; W. Fenical, Appl. Environ. Microbiol. 2002, 68, 5005–5011. R. H. Feling; G. O. Buchanan; T. J. Mincer; C. A. Kauffman; P. R. Jensen; W. Fenical, Angew. Chem. Int. Ed. 2003, 42, 355–357. H. C. Kwon; C. A. Kauffman; P. R. Jensen; W. Fenical, J. Am. Chem. Soc. 2006, 128, 1622–1632. P. G. Williams; R. N. Asolkar; T. Kondratyuk; J. M. Pezzuto; P. R. Jensen; W. Fenical, J. Nat. Prod. 2007, 70, 83–88. D. C. Oh; W. K. Strangman; C. A. Kauffman; P. R. Jensen; W. Fenical, Org. Lett. 2007, 9, 1525–1528. J. Piel, Nat. Prod. Rep. 2004, 21, 519–538. T.-W. Yu; H. G. Floss, Ansamitocins (Maytansanoids). In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 321–337. J. Piel; D. Butzke; N. Fusetani; D. Hui; M. Platzer; G. Wen; S. Matsunaga, J. Nat. Prod. 2005, 68, 472–479. J. Piel; I. Hofer; D. Hui, J. Bacteriol. 2004, 186, 1280–1286. J. Piel; D. Hui; G. Wen; D. Butzke; M. Platzer; N. Fusetani; S. Matsunaga, Proc. Nat. Acad. Sci. U.S.A. 2004, 101, 16222–16227. L. P. Partida-Martinez; C. Hertweck, Nature 2005, 437, 884–888. D. Downes; S. A. Laird; C. Klein; B. K. Carney, Biodiversity Prospecting Contract. In Biodiversity Prospecting: Using Genetic Resources for Sustainable Development, W. V. Reid, S. A. Laird, C. A. Meyer, R. Gamez, A. Sittenfeld, D. H. Janzen, M. A. Gollin, C. Juma, Eds.; World Resources Institute: Washington, DC, 1993; pp 255–287. K. ten Kate; A. Wells, The access and benefit-sharing policies of the United States National Cancer Institute: A comparative account of the discovery and development of the drugs Calanolide and Topotecan. Benefit-Sharing Case Study. Submission to the Executive Secretary of the Convention on Biological Diversity. Case study http://www.cbd.int K. ten Kate; S. A. Laird, The Commercial Use of Biodiversity. Access to Genetic Resources and Benefit-Sharing; Earthscan Publications Ltd.: London, UK, 1999. J. P. Rosenthal, Pharm. Biol. 1999, 37, 5. M. J. Balick, Ethnobotany and the Identification of Therapeutic Agents from the Rainforest. In Bioactive Compounds from Plants; D. J. Chadwick; J. Marsh, Eds.; Wiley: Chichester, 1990; pp 22–39. P. A. Cox, Ethnopharmacology and the Search for New Drugs. In Bioactive Compounds from Plants; D. J. Chadwick, J. Marsh, Eds.; Wiley: Chichester, 1990; pp 40–55. N. R. Farnsworth, The Role of Ethnopharmacology in Drug Development. In Bioactive Compounds from Plants; D. J. Chadwick, J. Marsh, Eds.; Wiley: Chichester, 1990; pp 2–21. C. Khosla, J. Org. Chem. 2000, 65, 8127–8133. J. Staunton; K. J. Weissman, Nat. Prod. Rep. 2001, 18, 380–416. C. T. Walsh, Science 2004, 303, 1805–1810. C. T. Walsh, Acc. Chem. Res. 2007, 41, 4–10. J. Clardy; C. T. Walsh, Nature 2004, 432, 829–837. 170 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 108. M. G. Thomas; K. A. Bixby; B. Shen, Combinatorial Biosynthesis of Anticancer Natural Products. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 519–552. 109. B. Julien; S. Shah; R. Ziemann; R. Goldman; L. Katz; C. Khosla, Gene 2000, 249, 153–160. 110. I. Molnar; T. Schupp; M. Ono; R. E. Zirkle; M. Milnamow; B. Nowak-Thompson; N. Engel; C. Toupet; A. Stratmann; D. D. Cyr; J. Gorlach; J. M. Mayo; A. Hu; S. Goff; J. Schmid; J. M. Ligon, Chem. Biol. 2000, 7, 97–109. 111. J. Lau; S. Frykman; R. Regentin; S. Ou; H. Tsuruta; P. Licari, Biotechnol. Bioeng. 2002, 78, 280–288. 112. K. C. Nicolaou; D. Vourloumis; N. Winssinger; P. S. Baran, Angew. Chem. Int. Ed. 2000, 39, 44–122. 113. K. C. Nicolaou; S. A. Snyder, Angew. Chem. Int. Ed. 2005, 44, 1012–1044. 114. M. Freemantle, Chem. & Eng. News 2004, 82 (9), 33–35. 115. S. J. Mickel; D. Niederer; R. Daeffler; A. Osmani; E. Kuesters; E. Schmid; K. Schaer; R. Gamboni; W. C. Chen; E. Loeser; F. R. Kinder, Jr.; K. Konigsberger; K. Prasad; T. M. Ramsey; O. Repic; R. M. Wang; G. Florence; I. Lyothier; I. Paterson, Org. Process Res. Dev. 2004, 8, 122–130. 116. M. J. Yu; Y. Kishi; B. A. Littlefield, Discovery of E7389, a Fully Synthetic Macrocyclic Ketone Analog of Halichondrin B. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 241–265. 117. E. Flahive; J. Srirangam, The Dolastatins: Novel Antitumor Agents from Dolabella auricularia. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 191–214. 118. S. Borman, Chem. Eng. News 2003, 81 (43), 45–56. 119. S. Class, Chem. Eng. News 2002, 80 (48), 39–49. 120. S. Borman, Chem. Eng. News 2004, 82 (40), 32–40. 121. M. D. Burke; E. M. Berger; S. L. Schreiber, Science 2003, 302, 613–618. 122. M. D. Burke; S. L. Schreiber, Angew. Chem. Int. Ed. 2004, 43, 46–58. 123. M. Thutewohl; L. Kissau; B. Popkirova; I.-M. Karaguni; T. Nowak; M. Bate; J. Kuhlmann; O. Muller; H. Waldmann, Angew. Chem. Int. Ed. 2002, 41, 3616–3620. 124. P. Stahl; L. Kissau; R. Mazitschek; A. Giannis; H. Waldmann, Angew. Chem. Int. Ed. 2002, 41, 1174–1178. 125. D. Brohm; S. Metzger; A. Bhargava; O. Muller; F. Lieb; H. Waldmann, Angew. Chem. Int. Ed. 2002, 41, 307–311. 126. R. Breinbauer; I. R. Vetter; H. Waldmann, Angew. Chem. Int. Ed. 2002, 41, 2878–2890. 127. R. Breinbauer; M. Manger; M. Scheck; H. Waldmann, Curr. Med. Chem. 2002, 9, 2129–2145. 128. A. Ganesan, Curr. Opin. Biotech. 2004, 15, 584–590. 129. M. A. Koch; H. Waldmann, Drug Discov. Today 2005, 10, 471–483. 130. K. C. Nicolaou; S. Kim; J. A. Pfefferkorn; J. Xu; T. Oshima; S. Hosokawa; T. Li, Angew. Chem. Int. Ed. 1998, 37, 1418–1421. 131. B. E. Evans; K. E. Rittle; M. G. Bock; R. M. DiPardo; R. M. Fredinger; W. L. Whitter; G. F. Lundell; D. F. Veber; P. S. Anderson; R. S. L. Chang; V. J. Lotti; D. J. Cerino; T. B. Chen; P. Kling; K. A. Kunkel; J. P. Springer; J. Hirshfield, J. Med. Chem. 1988, 31, 2235–2246. 132. K. C. Nicolaou; J. A. Pfefferkorn; A. J. Roecker; G. Q. Cao; S. Barluenga; H. J. Mitchell, J. Am. Chem. Soc. 2000, 122, 9939–9953. 133. K. C. Nicolaou; J. A. Pfefferkorn; S. Barluenga; H. J. Mitchell; A. J. Roecker; G. Q. Cao, J. Am. Chem. Soc. 2000, 122, 9968–9976. 134. K. C. Nicolaou; J. A. Pfefferkorn; H. J. Mitchell; A. J. Roecker; S. Barluenga; G. Q. Cao; R. L. Affleck; J. E. Lillig, J. Am. Chem. Soc. 2000, 122, 9954–9967. 135. L. Meijer, Oncologie 2003, 5, 311–326. 136. D. J. Newman; G. M. Cragg; K. M. Snader, Nat. Prod. Rep. 2000, 17, 215–234. 137. J. Wang; S. M. Soisson; K. Young; W. Shoop; S. Kodali; A. Galgoci; R. Painter; G. Parthasarathy; Y. S. Tang; R. Cummings; S. Ha; K. Dorso; M. Motyl; H. Jayasuriya; J. Ondeyka; K. Herath; C. Zhang; L. Hernandez; J. Allocco; A. Basilio; J. R. Tormo; O. Genilloud; F. Vicente; F. Pelaez; L. Colwell; S. H. Lee; B. Michael; T. Felcetto; C. Gill; L. L. Silver; J. D. Hermes; K. Bartizal; J. Barrett; D. Schmatz; J. W. Becker; D. Cully; S. B. Singh, Nature 2006, 441, 358–361. 138. J.-F. Tremblay, Chem. Eng. News 2006, 84 (10), 44. 139. J. D. Coombes, New Drugs from Natural Sources; IBC Technical Services: London, 1992. 140. J. Klekota; E. Brauner; F. P. Roth; S. L. Schreiber, J. Chem. Inf. Model. 2006, 46, 1549–1562. 141. B. Liu; S. Li; J. Hu, Am. J. Pharmacogen. 2004, 4, 263–276. 142. R. Mullin, Chem. Eng. News 2004, 82 (30), 23–32. 143. M. R. Boyd; K. D. Paull, Drug Dev. Res. 1995, 34, 91–109. 144. R. K. Johnson; H. F. Bartus; G. A. Hofmann; J. O. Bartus; S.-M. Mong; L. F. Faucette; F. L. McCabe; J. A. Chan; C. K. Mirabelli, Discovery of New DNA-Reactive Drugs. In In Vitro and In Vivo Models for Detection of New Antitumor Drugs; L. J. Hanka, T. Kondo, R. J. White, Eds.; University of Tokyo Press: Tokyo, 1986; pp 15–26. 145. B. N. Meyer; N. R. Ferrigni; J. E. Putnam; L. B. Jacobsen; D. E. Nichols; J. L. McLaughlin, Planta Med. 1982, 45, 31–34. 146. J. L. McLaughlin, Crown Gall Tumors on Potato Discs and Brine Shrimp Lethality: Two simple Bioassays for Higher Plant Screening and Fractionation. In Methods in Plant Biochemistry: Assays for Bioactivity; K. Hostettmann, Ed.; Academic Press: San Diego, 1991; Vol. 6, pp 1–32. 147. M. G. Hollingshead; M. C. Alley; R. F. Camalier; B. J. Abbott; J. G. Mayo; L. Malspeis; M. R. Grever, Life Sci. 1995, 57, 131–141. 148. M. C. Alley; M. G. Hollingshead; D. J. Dykes; W. R. Waud, Human Tumor Xenograft Models in NCI Drug Development. In Cancer Drug Discovery and Development: Anticancer Drug Development Guide: Preclinical Screening, Clinical Trials, and Approval B. A. Teicher,P. A. Andrews, Eds.; Humana Press Inc.: Totowa, NJ, 2004; Chapter 7. 149. G. M. Cragg; D. J. Newman, J. Nat. Prod. 2004, 67, 232–244. 150. G. M. Cragg; D. G. I. Kingston; D. J. Newman, Eds., Anticancer Agents from Natural Products; Taylor & Francis Group: Boca Raton, FL, 2005. 151. D. G. I. Kingston, Taxol and Its Analogs. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 89–122. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 171 152. G. Hofle; H. Reichenbach, Epothilone, a Myxobacterial Metabolite with Promising Antitumor Activity. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 413–450. 153. S. P. Gunasekera; A. E. Wright, Chemistry and Biology of the Discodermolides, Potent Mitotic Spindle Poisons. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 171–190. 154. K. G. Pinney; C. Jelinek; K. Edvardsen; D. J. Chaplin; G. R. Pettit, The Discovery and Development of the Combretastatins. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 23–46. 155. F. Gueritte; J. Fahy, The Vinca Alkaloids. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 123–136. 156. R. J. Andersen; M. Roberge, HTI-286. A Synthetic Analog of the Antimitotic Natural Product Hemiasterlin. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 267–280. 157. D. J. Newman; G. M. Cragg, Curr. Drug Targets 2006, 7, 279–304. 158. M. Kondoh; T. Usui; S. Kobayashi; K. Tsuchiya; K. Nishikawa; T. Nishikiori; T. Mayumi; H. Osada, Cancer Lett. 1998, 126, 29–32. 159. T. Usui; H. Watanabe; Y. Tada; N. Kanoh; M. Kondoh; T. Asao; K. Takio; H. Watanabe; K. Nishikawa; T. Kitahara; H. Osada, Chem. Biol. 2004, 11, 799–806. 160. G. M. Cragg; D. J. Newman, J. Nat. Prod. 2004, 67, 232–244. 161. N. J. Rahier; C. J. Thomas; S. M. Hecht, Camptothecin and Its Analogs. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 5–22. 162. M. Prudhomme, Staurosporines and Structurally Related Indolocarbazoles as Antitumor Agents. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 499–518. 163. F. M. Arcamone, Anthracyclines. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 299–320. 164. K.-H. Lee; Z. Xiao, Podophyllotoxins and Analogs. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 71–88. 165. W. A. Denny, Expert Opin. Emerg. Drugs 2004, 9, 105–133. 166. K. M. Marshall; J. A. Holden; A. Koller; Y. Kashman; B. R. Copp; L. R. Barrows, Anti-Cancer Drugs 2004, 15, 907–913. 167. K. M. Marshall; L. R. Barrows, Nat. Prod. Rep. 2004, 21, 731–751. 168. M. Yoshida; R. Furumai; M. Nishiyama; Y. Komatsu; N. Nishino; S. Horinouchi, Cancer Chemother. Pharmacol. 2001, 48, S20–S26. 169. S. W. Remiszewski, Curr. Opin. Drug. Discov. Devel. 2002, 5, 487–499. 170. M. Yoshida; A. Matsuyama; Y. Komatsu; N. Nishino, Curr. Med. Chem. 2003, 10, 2351–2358. 171. N. Tsuji; N. Kobayashi; K. Nagashima; Y. Wakisaka; K. Koizumi, J. Antibiot. 1976, 29, 1–6. 172. T. Vanhaecke; P. Papeleu; G. Elaut; V. Rogiers, Curr. Med. Chem. 2004, 11, 1629–1643. 173. H. Brinkmann; A. L. Dahler; C. Popa; M. M. Sereweko; P. G. Parsons; B. G. Gabrielli; A. J. Burgess; N. A. Saunders, J. Biol. Chem. 2001, 276, 22491–22499. 174. J. P. Secrist; X. Zhou; V. M. Richon, Curr. Opin. Investig. Drugs 2003, 4, 1422–1427. 175. M. Jung; G. Brosch; D. Kolle; H. Scherf; C. Gerhauser; P. Loidl, J. Med. Chem. 1999, 42, 4669–4679. 176. S. W. Remiszewski; L. C. Sambucetti; P. Atadja; K. W. Bair; W. D. Cornell; M. A. Green; W. D. Cornell; M. A. Green; K. L. Howell; M. Jung; P. Kwon; N. Trogani; H. Walker, J. Med. Chem. 2002, 45, 753–757. 177. M. Kijima; M. Yoshida; K. Sugita; S. Horinouchi; T. Beppu, J. Biol. Chem. 1993, 268, 22429–22435. 178. Y. F. Hallock; G. M. Cragg, Pharm. Biol. 2003, 41 (Suppl.), 78–91. 179. P. Crews; W. H. Gerwick; F. J. Schmitz; D. France; K. W. Bair; A. E. Wright; Y. Hallock, Pharm. Biol. 2003, 41 (Suppl.), 39–52. 180. S. W. Remiszewski, Curr. Med. Chem. 2003, 10, 2393–2402. 181. V. Sandor; A. Senderowicz; S. Mertins; D. Sackett; E. Sausville; M. V. Blagosklonny; S. E. Bates, Br. J. Cancer 2000, 83, 817–825. 182. D. J. Newman; G. M. Cragg; K. M. Snader, J. Nat. Prod. 2003, 66, 1022–1037. 183. M. C. Christian; J. M. Pluda; T. C. Ho; S. G. Arbuck; A. J. Murgo; E. A. Sausville, Semin. Oncol. 1997, 24, 219–240. 184. P. M. Fischer; A. Gianella-Borradori, Expert Opin. Investig. Drugs 2005, 14, 457–477. 185. F. Mayer; S. Mueller; E. Malenke; M. Kuczyk; J. T. Hartmann; C. Bokemeyer, Investig. New Drugs 2005, 23, 205–211. 186. M. Suffness; D. J. Newman; K. M. Snader, Dicovery and Development of Antineoplastic Agents from Natural Sources. In Bioorganic Marine Chemistry; P. Scheuer, Ed.; Springer-Verlag: Berlin-Heidelberg, 1989; Vol. 3, pp 131–167. 187. G. R. Pettit, The Bryostatins. In Progress in the Chemistry of Organic Natural Products; W. Hertz, G. W. Kirby, W. Steglich, C. Tamm, Eds.; Springer-Verlag: New York, 1991; Vol. 57, pp 153–195. 188. G. R. Pettit, J. Nat. Prod. 1996, 59, 812–821. 189. D. J. Newman, Bryostatin – from Bryozoan to Cancer Drug. In Bryozoans in Space and Time; D. P. Gordon, A. M. Smith, J. A. Grant-Mackie, Eds.; NIWA: Wellington, NZ, 1996; pp 9–17. 190. R. Mutter; M. Wills, Bioorg. Med. Chem. 2000, 8, 1841–1860. 191. G. R. Pettit; C. L. Herald; F. Hogan, Biosynthetic Products for Anticancer Drug Design and Treatment: The Bryostatins. In Anticancer Drug Development; B. C. Baguley, D. J. Kerr, Eds.; Academic Press: San Diego, 2002; pp 203–235. 192. K. J. Hale; M. C. Hummersone; S. Manaviazar; M. Frigerio, Nat. Prod. Rep. 2002, 19, 413–453. 193. D. J. Newman, The Bryostatins. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 137–150. 194. M. Kageyama; T. Tamura; M. H. Nantz; J. C. Roberts; P. Somfai; D. C. Whritenour; S. Masamune, J. Am. Chem. Soc. 1990, 112, 7407–7408. 195. D. A. Evans; P. H. Carter; E. M. Carreira; A. B. Charette; J. A. Prunet; M. Lautens, J. Am. Chem. Soc. 1999, 121, 7540–7552. 196. K. Ohmori; Y. Ogawa; T. Obitsu; Y. Ishikawa; S. Nishiyama; S. Yamamura, Angew. Chem. Int. Ed. 2000, 39, 2290–2294. 197. R. D. Norcross; I. Paterson, Chem. Rev. 1995, 95, 2041–2114. 198. P. A. Wender; K. F. Koehler; N. A. Sharkey; M. L. Dell’Aquila; P. M. Blumberg, Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 4214–4218. 172 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 199. P. A. Wender; C. M. Cribbs; K. F. Koehler; N. A. Sharkey; C. L. Herald; Y. Kamano; G. R. Pettit; P. M. Blumberg, Proc. Natl. Acad. Sci. U.S.A. 1988, 85, 7917 –7201. 200. P. A. Wender; J. De Brabander; P. G. Harran; K. W. Hinkle; B. Lippa; G. R. Pettit, Tetrahedron Lett. 1998, 39, 8625–8628. 201. P. A. Wender; J. De Brabander; P. G. Harran; J.-M. Jimenez; M. T. F. Koehler; B. Lippa; C.-M. Park; M. Shiozaki, J. Am. Chem. Soc. 1998, 120, 4534–4535. 202. P. A. Wender; J. De Brabander; P. G. Harran; J.-M. Jimenez; M. T. F. Koehler; B. Lippa; C.-M. Park; C. Siedenbiedel; G. R. Pettit, Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6624–6629. 203. P. A. Wender; K. W. Hinkle; M. T. F. Koehler; B. Lippa, Med. Res. Rev. 1999, 19, 388–407. 204. P. A. Wender; B. Lippa, Tetrahedron Lett. 2000, 41, 1007–1011. 205. P. A. Wender; K. W. Hinkle, Tetrahedron Lett. 2000, 41, 6725–6729. 206. P. A. Wender; J. L. Baryza; C. E. Bennett; F. C. Bi; S. E. Brenner; M. O. Clarke; J. C. Horan; C. Kan; E. Lacote; B. Lippa; P. G. Nell; T. M. Turner, J. Am. Chem. Soc. 2002, 124, 13648–13649. 207. P. A. Wender; A. V. W. Mayweg; C. L. VanDeusen, Org. Lett. 2003, 5, 277–279. 208. P. A. Wender; M. T. F. Koehler; M. Sendzik, Org. Lett. 2003, 5, 4549–4552. 209. L. Meijer; E. Raymond, Acc. Chem. Res. 2003, 36, 417–425. 210. C. W. Parker; B. Entsch; D. Letham, Phytochemistry 1986, 25, 303–310. 211. Y. T. Chang; N. G. Gray; G. R. Rosania; D. P. Sutherlin; S. Kwon; T. C. Norman; R. Sarohia; M. Leost; L. Meijer; P. G. Schultz, Chem. Biol. 1999, 6, 361–375. 212. T. Maugard; E. Enaud; P. Choisy; M. D. Legoy, Phytochemistry 2001, 58, 897–904. 213. C. J. Cooksey, Molecules 2001, 6, 736–769. 214. I. A. MacNeil; C. L. Tiong; C. Minor; P. R. August; T. H. Grossman; K. A. Loiacono; B. A. Lynch; T. Phillips; S. Narula; R. Sundaramoorthi; A. Tyler; T. Aldredge; H. Long; M. Gilman; D. Holt; M. S. Osburne, J. Mol. Microbiol. Biotechnol. 2001, 3, 301–308. 215. J. Adachi; Y. Mori; S. Matsui; H. Takigam; J. Fujino; H. C. A. Kitagawa; C. H. Miller, III; T. Kato; K. Saeki; T. Matsuda, J. Biol. Chem. 2001, 276, 31475–31478. 216. Z. Xiao; Y. Hao; B. Liu; L. Qian, Leuk. Lymphoma 2002, 43, 1763–1768. 217. L. Meijer; A.-L. Skaltsounis; P. Magiatis; P. Polychronopoulos; M. Knockaert; M. Leost; X. P. Ryan; C. A. Vonica; A. Brivanlou; R. Dajani; C. Crovace; C. Tarricone; A. Musacchio; S. M. Roe; L. Pearl; P. Greengard, Chem. Biol. 2003, 10, 1255–1266. 218. P. Cohen; M. Goedert, Nat. Rev. Drug Discov. 2004, 3, 479–487. 219. M. Knockaert; M. Blondel; S. Bach; M. Leost; C. Elbi; G. L. Hager; S. R. Nagy; D. Han; M. Denison; M. Ffrench; X. P. Ryan; P. Magiatis; P. Polychronopoulos; P. Greengard; L. Skaltsounis; L. Meijer, Oncogene 2004, 23, 4400–4412. 220. M. S. Denison; S. R. Nagy, Ann. Rev. Pharmacol.Toxicol. 2003, 43, 309–334. 221. P. Polychronopoulos; P. Magiatis; A.-L. Skaltsounis; V. Myrianthopoulos; E. Mikros; A. Tarricone; A. Musacchio; S. M. Roe; L. Pearl; M. Leost; P. Greengard; L. Meijer, J. Med. Chem. 2004, 47, 935–946. 222. M. Gompel; M. Leost; E. B. De Kier Joffe; L. Puricelli; L. H. Franco; J. Palermo; L. Meijer, Bioorg. Med. Chem. Lett. 2004, 14, 1703–1707. 223. P. M. Fischer, Curr. Med. Chem. 2004, 11, 1563–1583. 224. E. V. Koonin; Y. I. Wolf; G. P. Karev, Nature 2003, 420, 218–223. 225. J. Kobayashi; T. Madono; H. Shigemori, Tetrahedron 1995, 51, 10867–10974. 226. B.-N. Zhou; R. K. Johnson; M. R. Mattern; P. W. Fisher; D. G. I. Kingston, Org. Lett. 2001, 3, 4047–4049. 227. L. D. Arnold; D. J. Calderwood; R. W. Dixon; D. N. Johnston; J. S. Kamens; R. Munschauer; P. Rafferty; S. E. Ratnofsky, Bioorg. Med. Chem. Lett. 2000, 10, 2167–2170. 228. A. F. Burchat; D. J. Calderwood; G. C. Hirst; N. J. Holman; D. N. Johnston; R. Munschauer; P. Rafferty; G. B. Tometzki, Bioorg. Med. Chem. Lett. 2000, 10, 2171–2174. 229. A. F. Burchat; D. J. Calderwood; M. M. Friedman; G. C. Hirst; B. Li; P. Rafferty; K. Ritter; B. S. Skinner, Bioorg. Med. Chem. Lett. 2002, 12, 1687–1690. 230. D. J. Calderwood; D. N. Johnston; R. Munschauer; P. Rafferty, Bioorg. Med. Chem. Lett. 2002, 12, 1683–1686. 231. L. Kissau; P. Stahl; R. Mazitschek; A. Giannis; H. Waldmann, J. Med. Chem. 2003, 46, 2917–2931. 232. M. A. Koch; L.-O. Wittenberg; S. Basu; D. A. Jeyaraj; E. Gourzouidou; K. Reinecke; A. Odermatt; H. Waldmann, Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16721–16726. 233. D. B. Solit; G. Chiosis, Drug Discov. Today 2008, 13, 38–43. 234. G. Chiosis; E. Caldas Lopes; D. Solit, Curr. Opin. Investig. Drugs 2006, 7, 534–541. 235. G. Chiosis; A. Rodina; K. Moulick, Anticancer Agents Med. Chem. 2006, 6, 1–8. 236. K. M. Snader, Benzoquinone Ansamycins. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 339–356. 237. J.-Y. Le Brazidec; A. Kamal; D. Busch; L. Thao; L. Zhang; G. Timony; R. Grecko; K. Trent; R. Lough; T. Salazar; S. Khan; F. Burrows; M. F. Boehm, J. Med. Chem. 2004, 47, 3865–3873. 238. Z.-Q. Tian; Y. Liu; D. Zhang; Z. Wang; S. D. Dong; C. W. Carreras; Y. Zhou; G. Rastelli; D. V. Santi; D. C. Myles, Bioorg. Med. Chem. 2004, 12, 5317–5329. 239. A. Kamal; L. Thao; J. Sensintaffar; L. Zhang; M. F. Boehm; L. C. Fritz; F. J. Burrows, Nature 2003, 425, 407–410. 240. Y.-S. Lee; M. G. Marcu; L. Neckers, Chem. Biol. 2004, 11, 991–996. 241. J. M. Jez; J. C. Chen; G. Rastelli; R. M. Stroud; D. V. Santi, Chem. Biol. 2003, 10, 361–368. 242. G. Chiosis; L. Neckers, ACS Chem. Biol. 2006, 1, 279–284. 243. G. Chiosis; M. N. Timaul; B. Lucas; P. N. Munster; F. F. Zheng; L. Sepp-Lorenzino; N. Rosen, Chem. Biol. 2001, 8, 289–299. 244. G. Chiosis; B. Lucas; H. Huezo; D. Solit; A. Basso; N. Rosen, Curr. Cancer Drug Targets 2003, 3, 371–376. 245. G. Chiosis, Curr. Top. Med. Chem. 2006, 6, 1183–1191. 246. G. Chiosis; H. Tao, IDrugs, 2006, 9, 778–782. 247. G. Chiosis; B. Lucas; A. Shtil; H. Huezo; N. Rosen, Bioorg. Med. Chem. 2002, 10, 3555–3564. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity 173 248. M. Vilenchik; D. Solit; A. Basso; H. Huezo; B. Lucas; H. He; N. Rosen; C. Spampinato; P. Modrich; G. Chiosis, Chem. Biol. 2004, 11, 787–797. 249. I. Llauger; H. He; J. Kim; J. Aguirre; N. Rosen; U. Peters; P. Davies; G. Chiosis, J. Med. Chem. 2005, 48, 2892–2905. 250. M. A. Biamonte; J. Shi; K. Hong; D. G. Hurst; I. Zhang; J. Fan; D. J. Busch; P. I. Karjian; A. A. Maldonado; J. L. Sensintaffar; Y. C. Yang; A. Kamal; R. E. Lough; K. Lundgren; F. J. Burrows; G. A. Timony; M. F. Boehm; S. R. Kasilbhatla, J. Med. Chem. 2006, 49, 817–828. 251. H. He; D. Zatorska; J. Kim; J. Aguirre; I. Llauger; Y. She; N. Wu; R. M. Immormino; D. T. Gewirth; G. Chiosis, J. Med. Chem. 2006, 49, 381–390. 252. Y. Ishii; S. Waxman; D. Germain, Anticancer Agents Med. Chem. 2007, 7, 359–365. 253. I. Zavrski; L. Kleeberg; M. Kaiser; U. Heider; J. Sterz; C. Jakob; O. Sezer, Curr. Pharm. Des. 2007, 13, 471–485. 254. W. S. Dalton, Semin. Oncol. 2004, 6 (Suppl. 16), 3–9. 255. A. F. Kisselev; A. L. Goldberg, Chem. Biol. 2001, 8, 739–758. 256. R. A. Kyle; S. V. Rajkumar, N. Engl. J. Med. 2004, 351, 1860–1873. 257. C. Montagut; A. Rovira; J. Albanell, Clin. Trans. Oncol. 2006, 8, 313–317. 258. S. V. Rajkumar; P. G. Richardson; T. Hideshima; K. C. Anderson, J. Clin. Oncol. 2005, 23, 630–639. 259. J. Adams, Cancer Cell 2003, 5, 417–421. 260. S. Omura; T. Fujimoto; K. Otoguro; K. Matsuzaki; R. Moriguchi; H. Tanaka; Y. Sasaki, J. Antibiot. 1991, 44, 113–116. 261. G. Fenteany; R. F. Standaert; W. S. Lane; S. Choi; E. J. Corey; S. L. Schreiber, Science 1995, 268, 726–731. 262. A. Craiu; M. Gaczynska; T. Akopian; C. F. Gramm; G. Fenteany; A. L. Goldberg; K. L. Rock, J. Biol. Chem. 1997, 272, 13437–13445. 263. L. R. Dick; A. A. Cruikshank; L. Grenier; F. D. Melandri; S. L. Nunes; R. L. Stein, J. Biol. Chem. 1996, 271, 7273–7276. 264. L. R. Dick; A. A. Cruikshank; A. T. Destree; L. Grenier; T. A. McCormack; F. D. Melandri; S. L. Nunes; V. J. Palombella; L. A. Parent; L. Plamodon; R. L. Stein, J. Biol. Chem. 1997, 272, 182–188. 265. E. J. Corey; W.-D. Z. Li, Chem. Pharm. Bull. (Tokyo) 1999, 47, 1–10. 266. E. J. Corey; W.-D. Z. Li; T. Nagamitsu; G. Fenteany, Tetrahedron 1999, 55, 3305–3316. 267. L. R. Reddy; P. Saravan; E. J. Corey, J. Am. Chem. Soc. 2004, 126, 6230–6231. 268. L. H. Meng; B. H. B. Kwok; N. Sin; C. M. Crews, Cancer Res. 1999, 59, 2798–2801. 269. L. H. Meng; R. Mohan; B. H. B. Kwok; M. Elofsson; N. Sin; C. M. Crews, Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 10403–10408. 270. Y. Koguchi; J. Kohno; M. Nishio; K. Takahashi; T. Okuda; T. Ohnuki; S. Komatsubara, J. Antibiot. 2000, 53, 105–109. 271. S. Nam; D. M. Smith; Q. P. Dou, J. Biol. Chem. 2001, 276, 13322–13330. 272. K. Rinehart; T. G. Holt; N. L. Fregeau; J. G. Stroh; P. A. Kiefer; F. Sun; L. H. Li; D. G. Martin, J. Org. Chem. 1990, 55, 4512–4515. 273. A. E. Wright; D. A. Forleo; G. P. Gunawardana; S. P. Gunasekera; F. E. Koehn; O. J. McConnell, J. Org. Chem. 1990, 55, 4508–4512. 274. C. van Kesteren; M. M. M. de Vooght; L. Lopez-Lazaro; R. A. A. Mathot; J. H. M. Schellens; J. M. Jimeno; J. H. Beijnen, Anti-Cancer Drugs 2003, 14, 487–502. 275. B. Dziegielewska; D. Kowalski; T. A. Beerman, Biochemistry 2004, 43, 14228–14237. 276. C. Cuevas; M. Perez; M. Martin; J. L. Chicharro; C. Fernandez-Rivas; M. Flores; A. Francesch; P. Gallefo; M. Zarzuelo; F. de la Calle; J. Garcia; C. Polanco; I. Rodriguez; I. Manzanares, Org. Lett. 2000, 2, 2545–2548. 277. R. Henriquez; G. Faircloth; C. Cuevas, Ecteinascidin 743 (ET-743; YondelisTM), Aplidin, and Kahalalide F. In Anticancer Agents from Natural Products; G. M. Cragg, D. G. I. Kingston, D. J. Newman, Eds.; Taylor & Francis: Boca Raton, FL, 2005; pp 215–240. 278. Anonymous, Yondelis recommended for E.U. approval in soft tissue sarcoma. DailyDrugNews.com (Daily Essentials) 23 July 2007. 279. Y. Li; X. Sun; J. T. LaMont; A. B. Pardee; C. J. Li, Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 2674–2678. 280. B. T. Choi; J. Cheong; Y. H. Choi, Anti-Cancer Drugs 2003, 14, 845–850. 281. A. G. Ravelo; A. Estevez-Braun; H. Chavez-Orellana; E. Perez-Sacau; D. Mesa-Siverio, Curr. Top. Med. Chem. 2004, 4, 241–265. 282. A. Giaccia; B. G. Siim; R. S. Johnson, Nat. Revs. Drug Discov. 2003, 2, 1–9. 283. M. W. Kunkel; D. L. Kirkpatrick; J. I. Johnson; G. Powis, Anticancer Drug. Des. 1997, 12, 659–670. 284. J. S. Lazo; K. Tamura; A. Vogt; J.-K. Jung; S. Rodriguez; R. Balachandran; B. W. Day; P. Wipf, J. Pharmacol. Exp. Ther. 2001, 296, 364–371. 285. P. Wipf; T. D. Hopkins; J.-K. Jung; S. Rodriguez; A. Birmingham; E. C. Southwick; J. S. Lazo; G. Powis, Bioorg. Med. Chem. Lett. 2001, 11, 2637–2641. 286. S. J. Welsh; R. R. Williams; A. Birmingham; D. J. Newman; D. L. Kirkpatrick; G. Powis, Mol. Can. Ther. 2003, 2, 235–243. 287. P. Wipf; S. M. Lynch; A. Birmingham; G. Tamayo; A. Jimenez; N. Campos; G. Powis, Org. Biomol. Chem. 2004, 2, 1651–1658. 288. R. Sakai; K. L. Rinehart; V. Kishore; B. Kundu; G. Faircloth; J. B. Gloer; J. R. Carney; M. Manikoshi; F. Sun; R. G. Hughes, Jr; D. Garcia-Gravalos; T. Garcia de Quesada; G. R. Wilson; R. M. Heid, J. Med. Chem. 1996, 39, 2819–2834. 289. C. Le Tourneau; E. Raymond; S. Faivre, Curr. Pharm. Des. 2007, 1, 397–403. 290. S. G. Gomez; J. A. Bueren; G. T. Faircloth; J. Jimeno; B. Albella, Exp. Hematol. 2003, 31, 1104–1111. 291. M. Vera; M. M. Joullie, Med. Res. Rev. 2002, 22, 102–145. 292. S. Kusari; M. Lamshöft; S. Zühlke; M. Spiteller, J. Nat. Prod. 2008, 71, 159–162. Web Release Date: 26 January 2008 (Communication). DOI: 10.1021/np070669k. 293. R. Balamurugan; F. J. Dekker; H. Waldmann, Mol. Biosyst. 2005, 1, 36–45. 174 Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity Biographical Sketches Gordon M. Cragg completed his undergraduate training in chemistry at Rhodes University, South Africa, and his D. Phil. (organic chemistry) from Oxford University in 1963. After 2 years of postdoctoral research at the University of California, Los Angeles, he returned to South Africa to join the Council for Scientific and Industrial Research. In 1966, he joined the chemistry department at the University of South Africa. He was transferred to the University of Cape Town in 1972. In 1979, he returned to the United States to join the Cancer Research Institute at Arizona State University. In 1985, he moved to the National Cancer Institute (NCI) in Bethesda, Maryland, and was appointed chief of the Natural Products Branch in 1989. He retired in December, 2004, and is currently serving as an NIH Special Volunteer. His major interests lie in the discovery of novel natural product agents for the treatment of cancer and AIDS, with an emphasis on multidisciplinary and international collaboration. He has given over 100 invited talks at conferences in many countries worldwide and has been awarded NIH Merit Awards for his contributions to the development of taxol (1991), leadership in establishing international collaborative research in biodiversity and natural products drug discovery (2004), and contributions to developing and teaching NIH technology transfer courses (2004). He was the president of the American Society of Pharmacognosy (1998–99) and was elected to honorary membership of the society in 2003. In November, 2006, he was awarded the William L. Brown Award for Plant Genetic Resources by the Missouri Botanical Garden at a 2-day symposium entitled ‘Realizing Nature’s Potential: The Once and Future King of Drug Discovery’ held in his honor. The Missouri Botanical Garden also named a recently discovered Madagascar plant in his honor, Ludia craggiana. He has established collaborations between the NCI and organizations in many countries promoting drug discovery from their natural resources. He has published over 150 papers related to these interests. David J. Newman is the current chief of the Natural Products Branch (NPB) in the Developmental Therapeutics Program at the National Cancer Institute in Frederick, MD. Nature as Source of Medicines; Novel Drugs from Nature; Screening for Antitumor Activity He was born in Grays, Essex, UK in 1939. In 1963, he received his M.Sc. in synthetic organic chemistry from the University of Liverpool working under Professor George Kenner, FRS, on pyrrole and porphyrin syntheses. Following time as a synthetic chemist at Ilford, Ltd., he joined the ARC’s Unit of Nitrogen Fixation at the University of London and then Sussex, as a research assistant in metallo-organic chemistry with Professor J. Chatt, FRS, transferring to the microbial biochemistry group in early 1966 as a graduate student under Professor John Postgate, FRS, and was awarded a D. Phil. in 1968 for the work on microbial electron transport proteins from Desulfovibrio. Following a move to the United States in September 1968, he did 2 years as a postdoc at the biochemistry department of the University of Georgia working on protein sequencing of Desulfovibrio ferredoxins, and then in 1970 joined SK&F in Philadelphia as a biological chemist. At SK&F, most work was related to biological chemistry and antibiotic discovery, and he left SK&F in 1985 when the antibiotic group was dissolved. For the next 6 years he worked in marine and microbial discovery programs (Air Products, SeaPharm, and Lederle) and then in 1991, joined the NPB as a chemist responsible for marine and microbial collection programs. He was given the NIH Merit Award in 2003 for this work and following Gordon Cragg’s retirement from the position of chief, NPB at the end of 2004, he was acting chief until appointed chief in late 2006. He has been the author or coauthor of over 110 papers, reviews, book chapters (and an editor, with Gordon Cragg and David Kingston of Anticancer Agents from Natural Products), and holds 18 patents. 175